8WM4
 
 | |
8WMC
 
 | |
8WMI
 
 | |
8WML
 
 | |
8JNK
 
 | |
8JNR
 
 | |
3ET2
 
 | |
5XXH
 
 | | Crystal Structure Analysis of the CBP | | 分子名称: | (3S)-1-[2-(3-ethanoylindol-1-yl)ethanoyl]piperidine-3-carboxylic acid, 1,2-ETHANEDIOL, CREB-binding protein, ... | | 著者 | Xiang, Q, Zhang, Y, Wang, C, Song, M. | | 登録日 | 2017-07-04 | | 公開日 | 2018-06-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Discovery and optimization of 1-(1H-indol-1-yl)ethanone derivatives as CBP/EP300 bromodomain inhibitors for the treatment of castration-resistant prostate cancer.
Eur J Med Chem, 147, 2018
|
|
5GUT
 
 | | The crystal structure of mouse DNMT1 (731-1602) mutant - N1248A | | 分子名称: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | 著者 | Chen, S.J, Ye, F. | | 登録日 | 2016-08-31 | | 公開日 | 2017-09-06 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Biochemical Studies and Molecular Dynamic Simulations Reveal the Molecular Basis of Conformational Changes in DNA Methyltransferase-1.
ACS Chem. Biol., 13, 2018
|
|
5Z1S
 
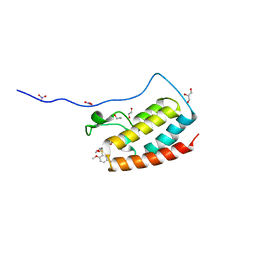 | | Crystal Structure Analysis of the BRD4(1) | | 分子名称: | 1,2-ETHANEDIOL, 5-bromo-2-methoxy-N-(6-methoxy-2,2-dimethyl-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-7-yl)benzene-1-sulfonamide, Bromodomain-containing protein 4, ... | | 著者 | Xu, Y, Zhang, Y, Xiang, Q, Song, M, Wang, C. | | 登録日 | 2017-12-28 | | 公開日 | 2019-01-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Y08060: A Selective BET Inhibitor for Treatment of Prostate Cancer.
Acs Med.Chem.Lett., 9, 2018
|
|
5GUV
 
 | |
5Z1R
 
 | | Crystal Structure Analysis of the BRD4 | | 分子名称: | 1,2-ETHANEDIOL, 5-bromo-N-(2,2-dimethyl-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-7-yl)-2-methoxybenzene-1-sulfonamide, Bromodomain-containing protein 4, ... | | 著者 | Xu, Y, Zhang, Y, Xiang, Q, Song, M, Wang, C. | | 登録日 | 2017-12-28 | | 公開日 | 2019-01-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.62 Å) | | 主引用文献 | Y08060: A Selective BET Inhibitor for Treatment of Prostate Cancer.
Acs Med.Chem.Lett., 9, 2018
|
|
5Z1T
 
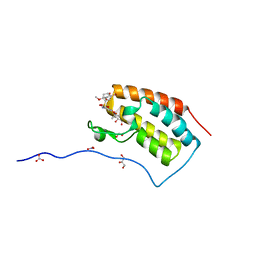 | | Crystal Structure Analysis of the BRD4(1) | | 分子名称: | 1,2-ETHANEDIOL, 5-bromo-N-(6-hydroxy-2,2-dimethyl-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-7-yl)-2-methoxybenzene-1-sulfonamide, Bromodomain-containing protein 4, ... | | 著者 | Xu, Y, Zhang, Y, Xiang, Q, Song, M, Wang, C. | | 登録日 | 2017-12-28 | | 公開日 | 2019-01-02 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.42 Å) | | 主引用文献 | Y08060: A Selective BET Inhibitor for Treatment of Prostate Cancer.
Acs Med.Chem.Lett., 9, 2018
|
|
8WKN
 
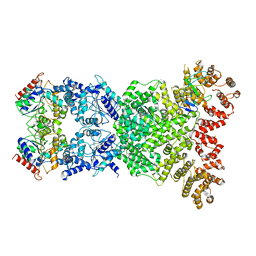 | | Cryo-EM structure of DSR2-DSAD1 | | 分子名称: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | 著者 | Zhang, H, Li, Z, Li, X.Z. | | 登録日 | 2023-09-28 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
8XKN
 
 | |
8W56
 
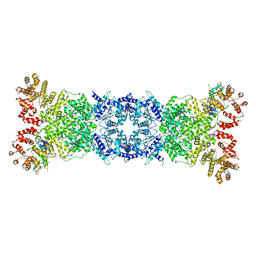 | | Cryo-EM structure of DSR2-DSAD1 state 1 | | 分子名称: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | 著者 | Zhang, H, Li, Z, Li, X.Z. | | 登録日 | 2023-08-25 | | 公開日 | 2024-05-01 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.59 Å) | | 主引用文献 | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
6AEI
 
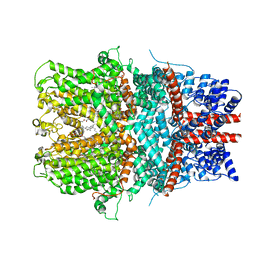 | | Cryo-EM structure of the receptor-activated TRPC5 ion channel | | 分子名称: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | 著者 | Duan, J, Li, Z, Li, J, Zhang, J. | | 登録日 | 2018-08-05 | | 公開日 | 2019-08-07 | | 最終更新日 | 2019-08-14 | | 実験手法 | ELECTRON MICROSCOPY (2.89 Å) | | 主引用文献 | Cryo-EM structure of TRPC5 at 2.8- angstrom resolution reveals unique and conserved structural elements essential for channel function.
Sci Adv, 5, 2019
|
|
7KN4
 
 | |
7KN3
 
 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain complexed with a pre-pandemic antibody S-B8 Fab | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, S-B8 Fab heavy chain, ... | | 著者 | Liu, H, Zhu, X, Wilson, I.A. | | 登録日 | 2020-11-04 | | 公開日 | 2021-09-22 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.251 Å) | | 主引用文献 | Neutralizing Antibodies to SARS-CoV-2 Selected from a Human Antibody Library Constructed Decades Ago.
Adv Sci, 9, 2022
|
|
3ET1
 
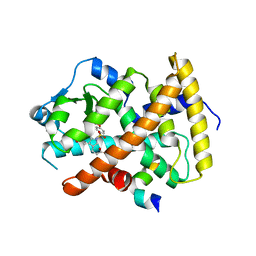 | |
3ET0
 
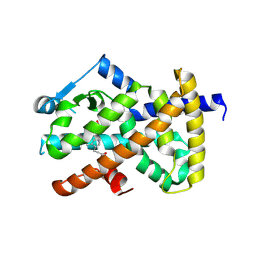 | |
3ET3
 
 | |
4W5I
 
 | | Crystal structure of human tankyrase 2 in complex with 1-methyl-7-phenyl-1,2,3,4,5,6-hexahydro-1,6- naphthyridin-5-one | | 分子名称: | 1-methyl-7-phenyl-2,3,4,6-tetrahydro-1,6-naphthyridin-5(1H)-one, GLYCEROL, SULFATE ION, ... | | 著者 | Haikarainen, T, Lehtio, L. | | 登録日 | 2014-08-18 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structure-based design, synthesis and evaluation in vitro of arylnaphthyridinones, arylpyridopyrimidinones and their tetrahydro derivatives as inhibitors of the tankyrases.
Bioorg.Med.Chem., 23, 2015
|
|
4UX4
 
 | | Crystal structure of human tankyrase 2 in complex with 1-methyl-7-(4- methylphenyl)-5-oxo-5,6-dihydro-1,6-naphthyridin-1-ium | | 分子名称: | (1S)-1-methyl-7-(4-methylphenyl)-5-oxo-1,5-dihydro-1,6-naphthyridin-1-ium, GLYCEROL, SULFATE ION, ... | | 著者 | Haikarainen, T, Lehtio, L. | | 登録日 | 2014-08-19 | | 公開日 | 2015-06-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-Based Design, Synthesis and Evaluation in Vitro of Arylnaphthyridinones, Arylpyridopyrimidinones and Their Tetrahydro Derivatives as Inhibitors of the Tankyrases.
Bioorg.Med.Chem., 23, 2015
|
|
7BZ5
 
 | | Structure of COVID-19 virus spike receptor-binding domain complexed with a neutralizing antibody | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of B38, Light chain of B38, ... | | 著者 | Wu, Y, Qi, J, Gao, F. | | 登録日 | 2020-04-26 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2.
Science, 368, 2020
|
|
