1VRK
 
 | |
3KY9
 
 | | Autoinhibited Vav1 | | 分子名称: | Proto-oncogene vav, ZINC ION | | 著者 | Tomchick, D.R, Rosen, M.K, Machius, M, Yu, B. | | 登録日 | 2009-12-04 | | 公開日 | 2010-02-23 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.731 Å) | | 主引用文献 | Structural and Energetic Mechanisms of Cooperative Autoinhibition and Activation of Vav1
Cell(Cambridge,Mass.), 140, 2010
|
|
8FOL
 
 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to SAM, alternate crystal form | | 分子名称: | CHLORIDE ION, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | 著者 | Moody, J.D, Saxton, A.J, Galambas, A, Lawrence, C.M, Broderick, J.B. | | 登録日 | 2022-12-31 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
8FO0
 
 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to a partially cleaved SAM molecule | | 分子名称: | IRON/SULFUR CLUSTER, POTASSIUM ION, Pyruvate formate-lyase 1-activating enzyme, ... | | 著者 | Moody, J.D, Saxton, A.J, Galambas, A, Lawrence, C.M, Broderick, J.B. | | 登録日 | 2022-12-29 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
8FSI
 
 | | The structure of a crystallizable variant of E. coli pyruvate formate-lyase activating enzyme bound to SAM | | 分子名称: | CHLORIDE ION, IRON/SULFUR CLUSTER, POTASSIUM ION, ... | | 著者 | Moody, J.D, Galambas, A, Lawrence, C.M, Broderick, J.B. | | 登録日 | 2023-01-10 | | 公開日 | 2023-05-24 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Computational engineering of previously crystallized pyruvate formate-lyase activating enzyme reveals insights into SAM binding and reductive cleavage.
J.Biol.Chem., 299, 2023
|
|
8FF2
 
 | |
8FF3
 
 | |
6XS6
 
 | | SARS-CoV-2 Spike D614G variant, minus RBD | | 分子名称: | Spike glycoprotein | | 著者 | Wang, X, Egri, S.B, Dudkina, N, Luban, J, Shen, K. | | 登録日 | 2020-07-15 | | 公開日 | 2020-07-22 | | 最終更新日 | 2020-11-11 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant.
Cell, 183, 2020
|
|
2HIO
 
 | | HISTONE OCTAMER (CHICKEN), CHROMOSOMAL PROTEIN | | 分子名称: | PROTEIN (HISTONE H2A), PROTEIN (HISTONE H2B), PROTEIN (HISTONE H3), ... | | 著者 | Arents, G, Moudrianakis, E.N. | | 登録日 | 1999-06-15 | | 公開日 | 2000-01-12 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
4UTB
 
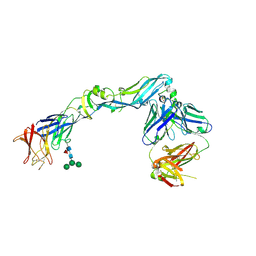 | | Crystal structure of dengue 2 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE2 A11 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11, ... | | 著者 | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | 登録日 | 2014-07-18 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (3.85 Å) | | 主引用文献 | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4V1V
 
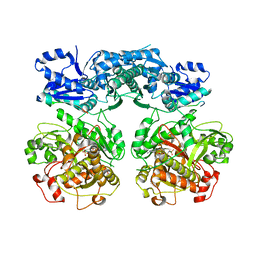 | |
4UT7
 
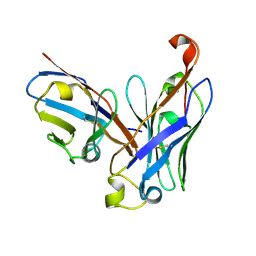 | | CRYSTAL STRUCTURE OF THE SCFV FRAGMENT OF THE BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 | | 分子名称: | BROADLY NEUTRALIZING HUMAN ANTIBODY EDE2 A11 | | 著者 | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | 登録日 | 2014-07-18 | | 公開日 | 2015-01-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
5XRW
 
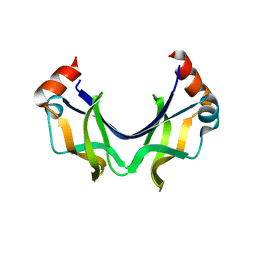 | |
5XYW
 
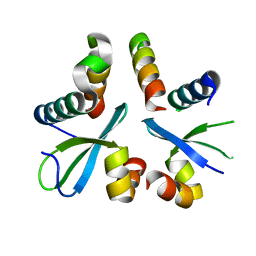 | |
5YA2
 
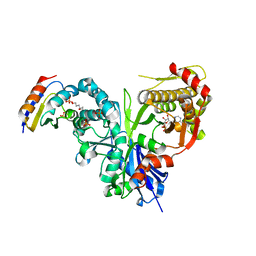 | | Crystal structure of LsrK-HPr complex with ADP | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Autoinducer-2 kinase, HEXANE-1,6-DIOL, ... | | 著者 | Ryu, K.S, Ha, J.H. | | 登録日 | 2017-08-30 | | 公開日 | 2018-07-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | Evidence of link between quorum sensing and sugar metabolism inEscherichia colirevealed via cocrystal structures of LsrK and HPr
Sci Adv, 4, 2018
|
|
4WSJ
 
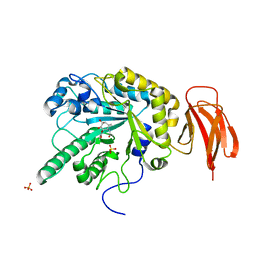 | | Crystal structure of a bacterial fucodiase in complex with 1-((1R,2R,3R,4R,5R,6R)-2,3,4-trihydroxy-5-methyl-7-azabicyclo[4.1.0]heptan-7-yl)ethan-1-one | | 分子名称: | Alpha-L-fucosidase, N-[(1S,2R,3R,4S,5R)-3,4,5-trihydroxy-2-methylcyclohexyl]acetamide, SULFATE ION | | 著者 | Davies, G.J, Wright, D.W. | | 登録日 | 2014-10-28 | | 公開日 | 2014-11-05 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.64 Å) | | 主引用文献 | In vitroandin vivocomparative and competitive activity-based protein profiling of GH29 alpha-l-fucosidases.
Chem Sci, 6, 2015
|
|
5YA1
 
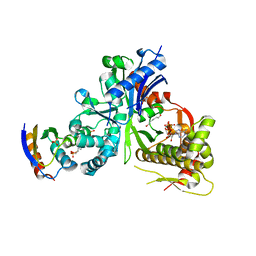 | | crystal structure of LsrK-HPr complex with ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Autoinducer-2 kinase, HEXANE-1,6-DIOL, ... | | 著者 | Ryu, K.S, Ha, J.H. | | 登録日 | 2017-08-29 | | 公開日 | 2018-07-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.701 Å) | | 主引用文献 | Evidence of link between quorum sensing and sugar metabolism inEscherichia colirevealed via cocrystal structures of LsrK and HPr
Sci Adv, 4, 2018
|
|
4WSK
 
 | | Crystal structure of a bacterial fucosidase with phenyl((1R,2R,3R,4R,5R,6R)-2,3,4-trihydroxy-5-methyl-7-azabicyclo[4.1.0]heptan-7-yl)methanone | | 分子名称: | Alpha-L-fucosidase, IMIDAZOLE, N-[(1S,2R,3R,4S,5R)-3,4,5-trihydroxy-2-methylcyclohexyl]benzamide, ... | | 著者 | Davies, G.J. | | 登録日 | 2014-10-28 | | 公開日 | 2014-11-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | In vitroandin vivocomparative and competitive activity-based protein profiling of GH29 alpha-l-fucosidases.
Chem Sci, 6, 2015
|
|
5XYV
 
 | |
5YA0
 
 | | Crystal structure of LsrK and HPr complex | | 分子名称: | Autoinducer-2 kinase, HEXANE-1,6-DIOL, PHOSPHATE ION, ... | | 著者 | Ryu, K.S, Ha, J.H. | | 登録日 | 2017-08-29 | | 公開日 | 2018-07-11 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.997 Å) | | 主引用文献 | Evidence of link between quorum sensing and sugar metabolism inEscherichia colirevealed via cocrystal structures of LsrK and HPr
Sci Adv, 4, 2018
|
|
5CZP
 
 | |
4X48
 
 | | Crystal structure of GluR2 ligand-binding core | | 分子名称: | GLUTAMIC ACID, Glutamate receptor 2, N-{(3S,4S)-4-[4-(5-cyanothiophen-2-yl)phenoxy]tetrahydrofuran-3-yl}propane-2-sulfonamide, ... | | 著者 | Pandit, J. | | 登録日 | 2014-12-02 | | 公開日 | 2015-05-06 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | The Discovery and Characterization of the alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptor Potentiator N-{(3S,4S)-4-[4-(5-Cyano-2-thienyl)phenoxy]tetrahydrofuran-3-yl}propane-2-sulfonamide (PF-04958242).
J.Med.Chem., 58, 2015
|
|
5DFE
 
 | |
3M5L
 
 | | Crystal structure of HCV NS3/4A protease in complex with ITMN-191 | | 分子名称: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, NS3/4A, SULFATE ION, ... | | 著者 | Schiffer, C.A, Romano, K.P. | | 登録日 | 2010-03-12 | | 公開日 | 2010-11-24 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (1.25 Å) | | 主引用文献 | Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V1U
 
 | |
