1IT7
 
 | | Crystal structure of archaeosine tRNA-guanine transglycosylase complexed with guanine | | 分子名称: | Archaeosine tRNA-guanine transglycosylase, GUANINE, MAGNESIUM ION, ... | | 著者 | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-01-11 | | 公開日 | 2002-05-22 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
5CH4
 
 | | Peptide-Bound State of Thermus thermophilus SecYEG | | 分子名称: | Protein translocase subunit SecE, Protein translocase subunit SecY, Putative preprotein translocase, ... | | 著者 | Tanaka, Y, Sugano, Y, Takemoto, M, Kusakizako, T, Kumazaki, K, Ishitani, R, Nureki, O, Tsukazaki, T. | | 登録日 | 2015-07-10 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.64 Å) | | 主引用文献 | Crystal Structures of SecYEG in Lipidic Cubic Phase Elucidate a Precise Resting and a Peptide-Bound State.
Cell Rep, 13, 2015
|
|
5B2Q
 
 | | Crystal structure of Francisella novicida Cas9 RHA in complex with sgRNA and target DNA (TGG PAM) | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Hirano, H, Nishimasu, H, Nakane, T, Ishitani, R, Nureki, O. | | 登録日 | 2016-02-01 | | 公開日 | 2016-03-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and Engineering of Francisella novicida Cas9
Cell, 164, 2016
|
|
5B43
 
 | | Crystal structure of Acidaminococcus sp. Cpf1 in complex with crRNA and target DNA | | 分子名称: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cpf1, DNA (34-MER), ... | | 著者 | Yamano, T, Nishimasu, H, Hirano, H, Nakane, T, Ishitani, R, Nureki, O. | | 登録日 | 2016-03-30 | | 公開日 | 2016-05-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of Cpf1 in Complex with Guide RNA and Target DNA
Cell, 165, 2016
|
|
5AWW
 
 | | Precise Resting State of Thermus thermophilus SecYEG | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Protein translocase subunit SecE, Protein translocase subunit SecY, ... | | 著者 | Tanaka, Y, Sugano, Y, Takemoto, M, Kusakizako, T, Kumazaki, K, Ishitani, R, Nureki, O, Tsukazaki, T. | | 登録日 | 2015-07-10 | | 公開日 | 2015-11-25 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.724 Å) | | 主引用文献 | Crystal Structures of SecYEG in Lipidic Cubic Phase Elucidate a Precise Resting and a Peptide-Bound State.
Cell Rep, 13, 2015
|
|
1IQ8
 
 | | Crystal Structure of archaeosine tRNA-guanine transglycosylase from Pyrococcus horikoshii | | 分子名称: | ARCHAEOSINE TRNA-GUANINE TRANSGLYCOSYLASE, MAGNESIUM ION, ZINC ION | | 著者 | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2001-07-09 | | 公開日 | 2002-05-22 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
1IU3
 
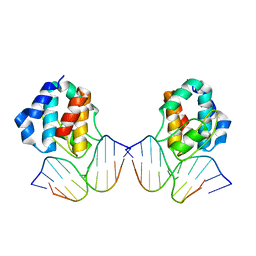 | | CRYSTAL STRUCTURE OF THE E.COLI SEQA PROTEIN COMPLEXED WITH HEMIMETHYLATED DNA | | 分子名称: | 5'-D(*AP*AP*GP*GP*AP*TP*CP*CP*AP*A)-3', 5'-D(*TP*TP*GP*GP*AP*TP*CP*CP*TP*T)-3', SeqA protein | | 著者 | Fujikawa, N, Kurumizaka, H, Nureki, O, Tanaka, Y, Yamazoe, M, Hiraga, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-02-26 | | 公開日 | 2003-06-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural and biochemical analyses of hemimethylated DNA binding by the SeqA protein.
Nucleic Acids Res., 32, 2004
|
|
1IVO
 
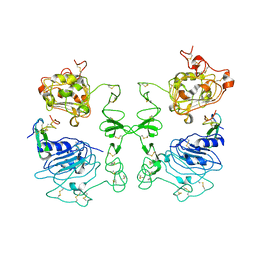 | | Crystal Structure of the Complex of Human Epidermal Growth Factor and Receptor Extracellular Domains. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal Growth Factor Receptor, ... | | 著者 | Ogiso, H, Ishitani, R, Nureki, O, Fukai, S, Yamanaka, M, Kim, J.H, Saito, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-03-28 | | 公開日 | 2002-10-16 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Crystal Structure of the Complex of Human Epidermal Growth Factor and Receptor Extracellular Domains.
Cell(Cambridge,Mass.), 110, 2002
|
|
5B2T
 
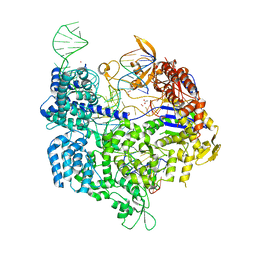 | | Crystal structure of the Streptococcus pyogenes Cas9 VRER variant in complex with sgRNA and target DNA (TGCG PAM) | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CRISPR-associated endonuclease Cas9, ... | | 著者 | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2016-02-02 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9
Mol.Cell, 61, 2016
|
|
5B2S
 
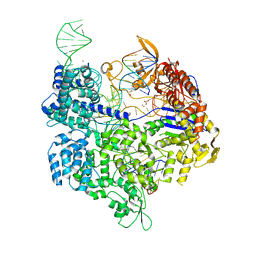 | | Crystal structure of the Streptococcus pyogenes Cas9 EQR variant in complex with sgRNA and target DNA (TGAG PAM) | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CRISPR-associated endonuclease Cas9, ... | | 著者 | Hirano, S, Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2016-02-02 | | 公開日 | 2016-03-23 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural Basis for the Altered PAM Specificities of Engineered CRISPR-Cas9
Mol.Cell, 61, 2016
|
|
5B2P
 
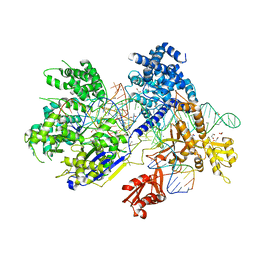 | | Crystal structure of Francisella novicida Cas9 in complex with sgRNA and target DNA (TGA PAM) | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | 著者 | Hirano, H, Nishimasu, H, Nakane, T, Ishitani, R, Nureki, O. | | 登録日 | 2016-02-01 | | 公開日 | 2016-03-02 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure and Engineering of Francisella novicida Cas9
Cell, 164, 2016
|
|
5D4I
 
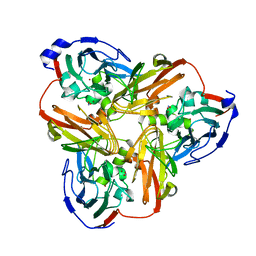 | | Intact nitrite complex of a copper nitrite reductase determined by serial femtosecond crystallography | | 分子名称: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | 著者 | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | 登録日 | 2015-08-07 | | 公開日 | 2016-03-09 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1J2B
 
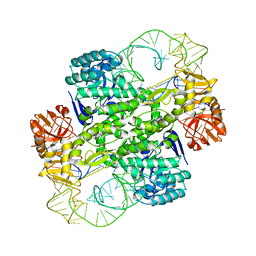 | | Crystal Structure Of Archaeosine tRNA-Guanine Transglycosylase Complexed With lambda-form tRNA(Val) | | 分子名称: | Archaeosine tRNA-guanine transglycosylase, MAGNESIUM ION, ZINC ION, ... | | 著者 | Ishitani, R, Nureki, O, Nameki, N, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-12-29 | | 公開日 | 2003-05-27 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Alternative Tertiary Structure of tRNA for Recognition by a Posttranscriptional Modification Enzyme
Cell(Cambridge,Mass.), 113, 2003
|
|
1IQ0
 
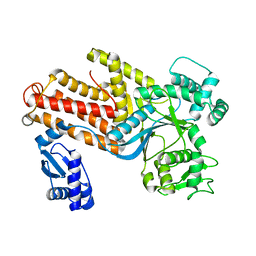 | | THERMUS THERMOPHILUS ARGINYL-TRNA SYNTHETASE | | 分子名称: | ARGINYL-TRNA SYNTHETASE | | 著者 | Shimada, A, Nureki, O, Goto, M, Takahashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2001-05-24 | | 公開日 | 2001-11-28 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural and mutational studies of the recognition of the arginine tRNA-specific major identity element, A20, by arginyl-tRNA synthetase.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1J1U
 
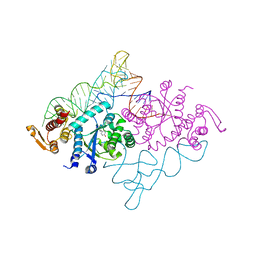 | | Crystal structure of archaeal tyrosyl-tRNA synthetase complexed with tRNA(Tyr) and L-tyrosine | | 分子名称: | MAGNESIUM ION, TYROSINE, Tyrosyl-tRNA synthetase, ... | | 著者 | Kobayashi, T, Nureki, O, Ishitani, R, Tukalo, M, Cusack, S, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-12-17 | | 公開日 | 2003-06-03 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structural basis for orthogonal tRNA specificities of tyrosyl-tRNA synthetases for genetic code expansion
NAT.STRUCT.BIOL., 10, 2003
|
|
7V93
 
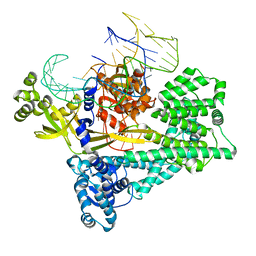 | | Cryo-EM structure of the Cas12c2-sgRNA binary complex | | 分子名称: | cas12c2, sgRNA | | 著者 | Kurihara, N, Hirano, H, Tomita, A, Kobayashi, K, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | 登録日 | 2021-08-24 | | 公開日 | 2022-04-13 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structure of the type V-C CRISPR-Cas effector enzyme.
Mol.Cell, 82, 2022
|
|
7V73
 
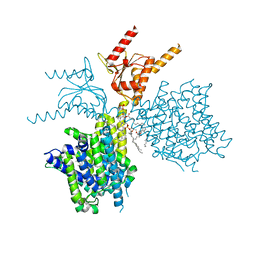 | | Thermostabilized human prestin in complex with chloride | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHLORIDE ION, CHOLESTEROL, ... | | 著者 | Futamata, H, Fukuda, M, Yamashita, K, Nishizawa, T, Nureki, O. | | 登録日 | 2021-08-21 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.52 Å) | | 主引用文献 | Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility.
Nat Commun, 13, 2022
|
|
7V75
 
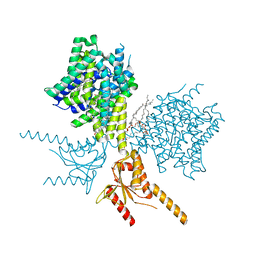 | | Thermostabilized human prestin in complex with salicylate | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-HYDROXYBENZOIC ACID, CHOLESTEROL, ... | | 著者 | Futamata, H, Fukuda, M, Yamashita, K, Nishizawa, T, Nureki, O. | | 登録日 | 2021-08-21 | | 公開日 | 2022-08-31 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.57 Å) | | 主引用文献 | Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility.
Nat Commun, 13, 2022
|
|
7V6B
 
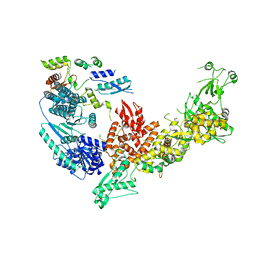 | | Structure of the Dicer-2-R2D2 heterodimer | | 分子名称: | Dicer-2, isoform A, R2D2 | | 著者 | Yamaguchi, S, Nishizawa, T, Kusakizako, T, Yamashita, K, Tomita, A, Hirano, H, Nishimasu, H, Nureki, O. | | 登録日 | 2021-08-20 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex.
Nature, 607, 2022
|
|
7V6C
 
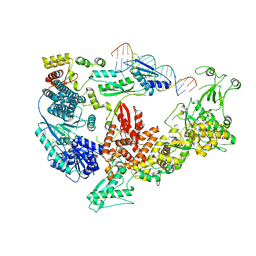 | | Structure of the Dicer-2-R2D2 heterodimer bound to small RNA duplex | | 分子名称: | Dicer-2, isoform A, R2D2, ... | | 著者 | Yamaguchi, S, Nishizawa, T, Kusakizako, T, Yamashita, K, Tomita, A, Hirano, H, Nishimasu, H, Nureki, O. | | 登録日 | 2021-08-20 | | 公開日 | 2022-03-23 | | 最終更新日 | 2024-06-12 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structure of the Dicer-2-R2D2 heterodimer bound to a small RNA duplex.
Nature, 607, 2022
|
|
3VVP
 
 | |
3UFZ
 
 | | Crystal structure of a Trp-less green fluorescent protein translated by the universal genetic code | | 分子名称: | Green fluorescent protein | | 著者 | Kawahara-Kobayashi, A, Araiso, Y, Matsuda, T, Yokoyama, S, Kigawa, T, Nureki, O, Kiga, D. | | 登録日 | 2011-11-02 | | 公開日 | 2012-10-17 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Simplification of the genetic code: restricted diversity of genetically encoded amino acids.
Nucleic Acids Res., 40, 2012
|
|
3VUW
 
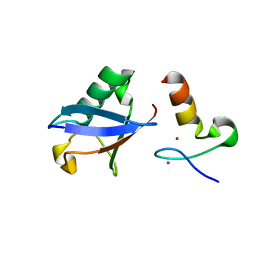 | | Crystal structure of A20 ZF7 in complex with linear ubiquitin, form I | | 分子名称: | POTASSIUM ION, Polyubiquitin-C, Tumor necrosis factor alpha-induced protein 3, ... | | 著者 | Nishimasu, H, Ishitani, R, Nureki, O. | | 登録日 | 2012-07-09 | | 公開日 | 2013-02-13 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-kappaB regulation
Embo J., 31, 2012
|
|
3VVR
 
 | | Crystal structure of MATE in complex with MaD5 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Putative uncharacterized protein, macrocyclic peptide | | 著者 | Tanaka, Y, Ishitani, R, Nureki, O. | | 登録日 | 2012-07-27 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Structural basis for the drug extrusion mechanism by a MATE multidrug transporter.
Nature, 496, 2013
|
|
3W1W
 
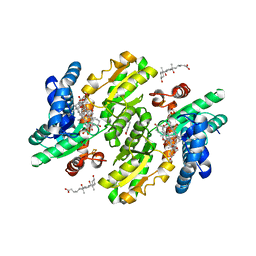 | | Protein-drug complex | | 分子名称: | 1,2-ETHANEDIOL, 2-HYDROXYBENZOIC ACID, CHOLIC ACID, ... | | 著者 | Ishii, R, Gupta, V, Yamaguchi, Y, Handa, H, Nureki, O. | | 登録日 | 2012-11-21 | | 公開日 | 2013-10-09 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.006 Å) | | 主引用文献 | Salicylic Acid induces mitochondrial injury by inhibiting ferrochelatase heme biosynthesis activity
Mol.Pharmacol., 84, 2013
|
|
