7MK3
 
 | | Crystal structure of NPR1 | | 分子名称: | CHLORIDE ION, GLYCEROL, Regulatory protein NPR1, ... | | 著者 | Cheng, J, Wu, Q, Zhou, P. | | 登録日 | 2021-04-21 | | 公開日 | 2022-03-16 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (3.06 Å) | | 主引用文献 | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
7MCE
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH 2-{(7P)-7-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-8-fluoro-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-3-yl}propan-2-ol | | 分子名称: | 2-{(7P)-7-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-8-fluoro-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-3-yl}propan-2-ol, Bromodomain-containing protein 4 | | 著者 | Sheriff, S. | | 登録日 | 2021-04-02 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Development of BET inhibitors as potential treatments for cancer: A new carboline chemotype.
Bioorg.Med.Chem.Lett., 51, 2021
|
|
8JYS
 
 | |
1I7E
 
 | | C-Terminal Domain Of Mouse Brain Tubby Protein bound to Phosphatidylinositol 4,5-bis-phosphate | | 分子名称: | L-ALPHA-GLYCEROPHOSPHO-D-MYO-INOSITOL-4,5-BIS-PHOSPHATE, TUBBY PROTEIN | | 著者 | Santagata, S, Boggon, T.J, Baird, C.L, Shan, W.S, Shapiro, L. | | 登録日 | 2001-03-08 | | 公開日 | 2001-06-27 | | 最終更新日 | 2023-08-09 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | G-protein signaling through tubby proteins.
Science, 292, 2001
|
|
7MK2
 
 | | CryoEM Structure of NPR1 | | 分子名称: | Regulatory protein NPR1, ZINC ION | | 著者 | Kumar, S, Zhou, Y, Dillard, L, Borgnia, M, Bartesaghi, A, Zhou, P. | | 登録日 | 2021-04-21 | | 公開日 | 2022-03-16 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural basis of NPR1 in activating plant immunity.
Nature, 605, 2022
|
|
8RC0
 
 | | Structure of the human 20S U5 snRNP | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Schneider, S, Galej, W.P. | | 登録日 | 2023-12-05 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structure of the human 20S U5 snRNP.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q91
 
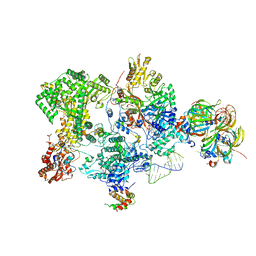 | | Structure of the human 20S U5 snRNP core | | 分子名称: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Schneider, S, Galej, W.P. | | 登録日 | 2023-08-19 | | 公開日 | 2024-03-27 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structure of the human 20S U5 snRNP.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5K8H
 
 | |
8WGV
 
 | | BA.2(S375) Spike (S6P)/hACE2 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | 著者 | Wei, X, Zhang, Z. | | 登録日 | 2023-09-22 | | 公開日 | 2024-02-28 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.92 Å) | | 主引用文献 | The receptor binding domain of SARS-CoV-2 Omicron subvariants targets Siglec-9 to decrease its immunogenicity by preventing macrophage phagocytosis.
Nat.Immunol., 25, 2024
|
|
2F5J
 
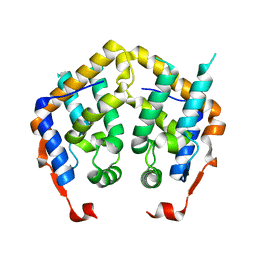 | |
4ONM
 
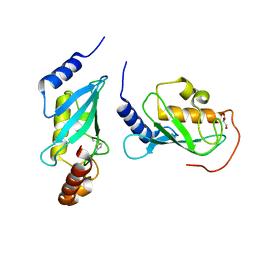 | | Crystal structure of human Mms2/Ubc13 - NSC697923 | | 分子名称: | 2-[(4-methylphenyl)sulfonyl]-5-nitrofuran, GLYCEROL, Ubiquitin-conjugating enzyme E2 N, ... | | 著者 | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | 登録日 | 2014-01-28 | | 公開日 | 2015-05-06 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Covalent Inhibition of Ubc13 Affects Ubiquitin Signaling and Reveals Active Site Elements Important for Targeting.
Acs Chem.Biol., 10, 2015
|
|
4ONN
 
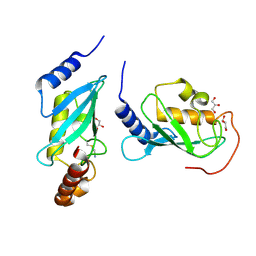 | | Crystal structure of human Mms2/Ubc13 - BAY 11-7082 | | 分子名称: | 3-[(4-methylphenyl)sulfonyl]prop-2-enenitrile, GLYCEROL, Ubiquitin-conjugating enzyme E2 N, ... | | 著者 | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | 登録日 | 2014-01-28 | | 公開日 | 2015-05-06 | | 最終更新日 | 2017-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Covalent Inhibition of Ubc13 Affects Ubiquitin Signaling and Reveals Active Site Elements Important for Targeting.
Acs Chem.Biol., 10, 2015
|
|
4OUS
 
 | |
4OUM
 
 | |
4ONL
 
 | | Crystal structure of human Mms2/Ubc13_D81N, R85S, A122V, N123P | | 分子名称: | Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 2 | | 著者 | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | 登録日 | 2014-01-28 | | 公開日 | 2015-05-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Covalent Inhibition of Ubc13 Affects Ubiquitin Signaling and Reveals Active Site Elements Important for Targeting.
Acs Chem.Biol., 10, 2015
|
|
4OUL
 
 | |
5YS1
 
 | | Crystal structure of Multicopper Oxidase CueO G304K mutant | | 分子名称: | Blue copper oxidase CueO, COPPER (II) ION | | 著者 | Wang, H.Q, Liu, X.Q, Zhao, J.T, Yue, Q.X, Yan, Y.H, Dong, Y.H, Fan, Y.L, Tian, J, Wu, N.F, Gong, Y. | | 登録日 | 2017-11-12 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.49 Å) | | 主引用文献 | Crystal structures of multicopper oxidase CueO G304K mutant: structural basis of the increased laccase activity
Sci Rep, 8, 2018
|
|
5YS5
 
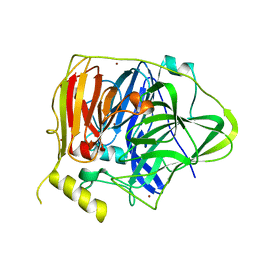 | | Crystal structure of Multicopper Oxidase CueO G304K mutant with seven copper ions | | 分子名称: | Blue copper oxidase CueO, COPPER (II) ION | | 著者 | Wang, H.Q, Liu, X.Q, Zhao, J.T, Yue, Q.X, Yan, Y.H, Dong, Y.H, Fan, Y.L, Tian, J, Wu, N.F, Gong, Y. | | 登録日 | 2017-11-13 | | 公開日 | 2018-10-17 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal structures of multicopper oxidase CueO G304K mutant: structural basis of the increased laccase activity
Sci Rep, 8, 2018
|
|
6KXW
 
 | | Crystal structure of human aquaporin AQP7 in bound to glycerol | | 分子名称: | Aquaporin-7, GLYCEROL | | 著者 | Zhang, L, Yao, D, Zhou, F, Zhang, Q, Zhou, L, Cao, Y. | | 登録日 | 2019-09-13 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.7 Å) | | 主引用文献 | The structural basis for glycerol permeation by human AQP7
Sci Bull (Beijing), 66, 2020
|
|
6LYC
 
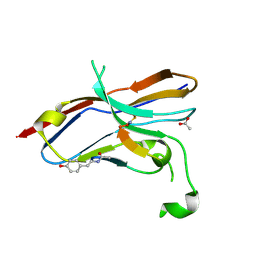 | | Crystal structure of the NOD SIRPa complex with D4-2 | | 分子名称: | ACETIC ACID, D4-2, SIRPa of the NOD mouse strain | | 著者 | Murata, Y, Matsuda, M, Nakagawa, A, Matozaki, T. | | 登録日 | 2020-02-14 | | 公開日 | 2020-07-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | Macrocyclic Peptide-Mediated Blockade of the CD47-SIRP alpha Interaction as a Potential Cancer Immunotherapy.
Cell Chem Biol, 27, 2020
|
|
7X83
 
 | | Cryo-EM structure of the TMEM106B fibril from normal elder | | 分子名称: | Transmembrane protein 106B | | 著者 | Xia, W.C, Zhao, Q.Y, Fan, Y, Sun, Y.P, Tao, Y.Q, Liu, C. | | 登録日 | 2022-03-11 | | 公開日 | 2022-06-15 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Generic amyloid fibrillation of TMEM106B in patient with Parkinson's disease dementia and normal elders.
Cell Res., 32, 2022
|
|
7X84
 
 | | Cryo-EM structure of the TMEM106B fibril from Parkinson's disease dementia | | 分子名称: | Transmembrane protein 106B | | 著者 | Zhao, Q.Y, Xia, W.C, Fan, Y, Sun, Y.P, Tao, Y.Q, Liu, C. | | 登録日 | 2022-03-11 | | 公開日 | 2022-06-15 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Generic amyloid fibrillation of TMEM106B in patient with Parkinson's disease dementia and normal elders.
Cell Res., 32, 2022
|
|
8K46
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K45
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nb4 nanobody, ... | | 著者 | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.66 Å) | | 主引用文献 | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
8K47
 
 | | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses including all major Omicron strains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Lu, Y, Gao, Y, Yao, H, Xu, W, Yang, H. | | 登録日 | 2023-07-17 | | 公開日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | A potent and broad-spectrum neutralizing nanobody for SARS-CoV-2 viruses, including all major Omicron strains.
MedComm (2020), 4, 2023
|
|
