7VGC
 
 | |
8IBQ
 
 | | Bromodomain and Extra-terminal Domain (BET) BRD4 | | 分子名称: | 7-[2-fluoranyl-3-(1,3,5-trimethylpyrazol-4-yl)phenyl]-1~{H}-imidazo[4,5-b]pyridine, Bromodomain-containing protein 4 | | 著者 | Cao, D, Zhiyan, D, Xiong, B. | | 登録日 | 2023-02-10 | | 公開日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Discovery of 1 H -Imidazo[4,5- b ]pyridine Derivatives as Potent and Selective BET Inhibitors for the Management of Neuropathic Pain.
J.Med.Chem., 66, 2023
|
|
8IDH
 
 | | Bromodomain and Extra-terminal Domain (BET) BRD4 | | 分子名称: | 7-[2-fluoranyl-3-(1,3,5-trimethylpyrazol-4-yl)phenyl]-1~{H}-imidazo[4,5-b]pyridine, Bromodomain-containing protein 4 | | 著者 | Cao, D, Zhiyan, D, Xiong, B. | | 登録日 | 2023-02-13 | | 公開日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Discovery of 1 H -Imidazo[4,5- b ]pyridine Derivatives as Potent and Selective BET Inhibitors for the Management of Neuropathic Pain.
J.Med.Chem., 66, 2023
|
|
7SQI
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C14-crypto Acyl Carrier Protein, AcpP | | 分子名称: | Acyl carrier protein, Beta-ketoacyl-ACP synthase I, N-{2-[(2Z)-3-chlorotetradec-2-enamido]ethyl}-N~3~-[(2R)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alaninamide, ... | | 著者 | Chen, A, Mindrebo, J.T, Davis, T.D, Noel, J.P, Burkart, M.D. | | 登録日 | 2021-11-05 | | 公開日 | 2022-08-03 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Mechanism-based cross-linking probes capture the Escherichia coli ketosynthase FabB in conformationally distinct catalytic states.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7SZ9
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C16:1-crypto Acyl Carrier Protein, AcpP | | 分子名称: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, Acyl carrier protein, N~3~-{(2R)-4-[(dihydroxyphosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-N-(2-{[(9Z)-hexadec-9-enoyl]amino}ethyl)-beta-alaninamide, ... | | 著者 | Mindrebo, J.T, Chen, A, Noel, J.P, Burkart, M.D. | | 登録日 | 2021-11-26 | | 公開日 | 2022-11-23 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Mechanism-based cross-linking probes capture the Escherichia coli ketosynthase FabB in conformationally distinct catalytic states.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
4M58
 
 | | Crystal Structure of an transition metal transporter | | 分子名称: | Cobalamin biosynthesis protein CbiM, NICKEL (II) ION | | 著者 | Yu, Y, Yan, C.Y, Zhang, B, Li, X.L, Gu, J.K. | | 登録日 | 2013-08-08 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.21 Å) | | 主引用文献 | Planar substrate-binding site dictates the specificity of ECF-type nickel/cobalt transporters
Cell Res., 24, 2014
|
|
4M5C
 
 | | Crystal Structure of an Truncated Transition metal Transporter | | 分子名称: | COBALT (II) ION, Cobalamin biosynthesis protein CbiM, HEXANE-1,6-DIOL | | 著者 | Yu, Y, Zhou, M.Z, Gu, J.K. | | 登録日 | 2013-08-08 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Planar substrate-binding site dictates the specificity of ECF-type nickel/cobalt transporters
Cell Res., 24, 2014
|
|
6KFW
 
 | |
5Z7C
 
 | |
6AIB
 
 | | Crystal structures of the N-terminal RecA-like domain 1 of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA | | 分子名称: | DEAD-box ATP-dependent RNA helicase CshA | | 著者 | Chengliang, W, Tian, T, Xiaobao, C, Xuan, Z, Jianye, Z. | | 登録日 | 2018-08-22 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6AIC
 
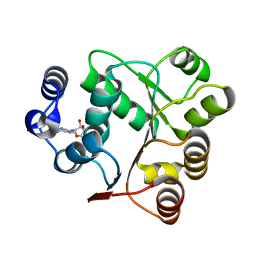 | | Crystal structures of the N-terminal domain of Staphylococcus aureus DEAD-box Cold shock RNA helicase CshA in complex with AMP | | 分子名称: | ADENOSINE MONOPHOSPHATE, DEAD-box ATP-dependent RNA helicase CshA | | 著者 | Tian, T, Chengliang, W, Xiaobao, C, Xuan, Z, Jianye, Z. | | 登録日 | 2018-08-22 | | 公開日 | 2018-11-21 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structures of the N-terminal domain of the Staphylococcus aureus DEAD-box RNA helicase CshA and its complex with AMP
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6M02
 
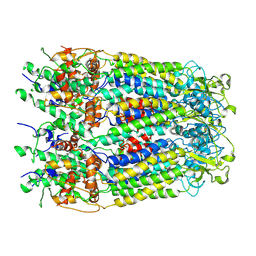 | | cryo-EM structure of human Pannexin 1 channel | | 分子名称: | Pannexin-1 | | 著者 | Ronggui, Q, Lili, D, Jilin, Z, Xuekui, Y, Lei, W, Shujia, Z. | | 登録日 | 2020-02-19 | | 公開日 | 2020-03-25 | | 最終更新日 | 2020-05-27 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Cryo-EM structure of human heptameric Pannexin 1 channel.
Cell Res., 30, 2020
|
|
2JRH
 
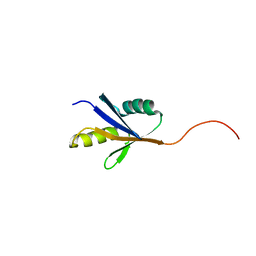 | |
7YHS
 
 | | Structure of Csy-AcrIF4-dsDNA | | 分子名称: | AcrIF4, CRISPR type I-F/YPEST-associated protein Csy2, CRISPR-associated protein Csy3, ... | | 著者 | Feng, Y, Zhang, L.X. | | 登録日 | 2022-07-14 | | 公開日 | 2023-03-29 | | 最終更新日 | 2024-07-03 | | 実験手法 | ELECTRON MICROSCOPY (3.37 Å) | | 主引用文献 | Anti-CRISPR protein AcrIF4 inhibits the type I-F CRISPR-Cas surveillance complex by blocking nuclease recruitment and DNA cleavage.
J.Biol.Chem., 298, 2022
|
|
7FAR
 
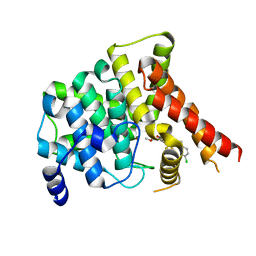 | | Crystal structure of PDE5A in complex with inhibitor L12 | | 分子名称: | 5-[bis(fluoranyl)methoxy]-2-[(4-chlorophenyl)methyl]-10-(3-methoxypropyl)-3,10-diazatricyclo[6.4.1.0^{4,13}]trideca-1,4(13),5,7-tetraen-9-one, MAGNESIUM ION, ZINC ION, ... | | 著者 | Wu, D, Huang, Y.Y, Luo, H.B. | | 登録日 | 2021-07-07 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.40006471 Å) | | 主引用文献 | Free energy perturbation (FEP)-guided scaffold hopping.
Acta Pharm Sin B, 12, 2022
|
|
7FAQ
 
 | | Crystal structure of PDE5A in complex with inhibitor L1 | | 分子名称: | 2-[(4-chlorophenyl)methyl]-5-methoxy-3,10-diazatricyclo[6.4.1.0^{4,13}]trideca-1,4(13),5,7-tetraen-9-one, MAGNESIUM ION, SULFATE ION, ... | | 著者 | Wu, D, Huang, Y.Y, Luo, H.B. | | 登録日 | 2021-07-07 | | 公開日 | 2022-05-18 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.200136 Å) | | 主引用文献 | Free energy perturbation (FEP)-guided scaffold hopping.
Acta Pharm Sin B, 12, 2022
|
|
8K62
 
 | | Crystal structure of ALKBH1 and 13h complex. | | 分子名称: | 1-[5-[[3-(trifluoromethyloxy)phenyl]methoxy]pyrimidin-2-yl]pyrazole-4-carboxylic acid, MANGANESE (II) ION, Nucleic acid dioxygenase ALKBH1 | | 著者 | Liang, X, Yinping, G, Feng, L, Jiang, Z, Ke, X, Shengyong, Y. | | 登録日 | 2023-07-24 | | 公開日 | 2024-07-31 | | 実験手法 | X-RAY DIFFRACTION (1.991 Å) | | 主引用文献 | Crystal structure of ALKBH1 and 13h complex
To Be Published
|
|
7EEJ
 
 | | Complex structure of glycoside hydrolase family 12 beta-1,3-1,4-glucanase with cellobiose | | 分子名称: | GLYCEROL, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose, glycoside hydrolase family 12 beta-1,3-1,4-glucanase | | 著者 | Jiang, Z.Q, Ma, J.W. | | 登録日 | 2021-03-18 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.47798049 Å) | | 主引用文献 | Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
J.Struct.Biol., 213, 2021
|
|
7EE2
 
 | | Structural insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.CQ31 | | 分子名称: | GLYCEROL, glycoside hydrolase family 12 beta-1,3-1,4-glucanase | | 著者 | Jiang, Z.Q, Ma, J. | | 登録日 | 2021-03-17 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.37011635 Å) | | 主引用文献 | Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
J.Struct.Biol., 213, 2021
|
|
7EEE
 
 | | Complex structure of glycoside hydrolase family 12 beta-1,3-1,4-glucanase with gentiobiose | | 分子名称: | GLYCEROL, beta-D-mannopyranose-(1-6)-beta-D-mannopyranose, glycoside hydrolase family 12 beta-1,3-1,4-glucanase | | 著者 | Jiang, Z.Q, Ma, J.W. | | 登録日 | 2021-03-18 | | 公開日 | 2022-03-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.660792 Å) | | 主引用文献 | Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
J.Struct.Biol., 213, 2021
|
|
7CBT
 
 | | The crystal structure of SARS-CoV-2 main protease in complex with GC376 | | 分子名称: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | 著者 | Shi, Y, Peng, G. | | 登録日 | 2020-06-13 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.346 Å) | | 主引用文献 | The preclinical inhibitor GS441524 in combination with GC376 efficaciously inhibited the proliferation of SARS-CoV-2 in the mouse respiratory tract.
Emerg Microbes Infect, 10, 2021
|
|
5XGH
 
 | | Crystal structure of PI3K complex with an inhibitor | | 分子名称: | 3-[(4-fluorophenyl)methylamino]-5-(4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenol, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | 著者 | Song, K, Yang, X, Zhao, Y, Jian, Z. | | 登録日 | 2017-04-13 | | 公開日 | 2018-04-25 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.97 Å) | | 主引用文献 | New Insights into PI3K Inhibitor Design using X-ray Structures of PI3K alpha Complexed with a Potent Lead Compound.
Sci Rep, 7, 2017
|
|
7C6P
 
 | | Bromodomain-containing 4 BD2 in complex with 3',4',7,8- Tetrahydroxyflavonoid | | 分子名称: | 2-[3,4-bis(oxidanyl)phenyl]-7,8-bis(oxidanyl)chromen-4-one, Bromodomain-containing protein 4 | | 著者 | Li, J, Yu, K, Luo, Y, Zheng, W, Liang, W, Zhu, J. | | 登録日 | 2020-05-22 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Discovery of the natural product 3',4',7,8-tetrahydroxyflavone as a novel and potent selective BRD4 bromodomain 2 inhibitor.
J Enzyme Inhib Med Chem, 36, 2021
|
|
7C2Z
 
 | | Bromodomain-containing 4 BD1 in complex with 3',4',7,8-Tetrahydroxyflavone | | 分子名称: | 2-[3,4-bis(oxidanyl)phenyl]-7,8-bis(oxidanyl)chromen-4-one, Bromodomain-containing protein 4, FORMIC ACID | | 著者 | Li, J, Zhu, J. | | 登録日 | 2020-05-10 | | 公開日 | 2021-05-12 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Discovery of the natural product 3',4',7,8-tetrahydroxyflavone as a novel and potent selective BRD4 bromodomain 2 inhibitor.
J Enzyme Inhib Med Chem, 36, 2021
|
|
7YGG
 
 | |
