4C7Q
 
 | |
8W6P
 
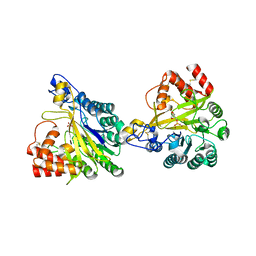 | | Crystal structure of dimeric murine SMPDL3A | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | 著者 | Zhang, C, Liu, P, Fan, S, Hou, Y. | | 登録日 | 2023-08-29 | | 公開日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | SMPDL3A is a cGAMP-degrading enzyme induced by LXR-mediated lipid metabolism to restrict cGAS-STING DNA sensing.
Immunity, 56, 2023
|
|
8W6R
 
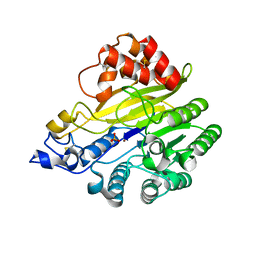 | | murine SMPDL3A bound to sulfate | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Acid sphingomyelinase-like phosphodiesterase 3a, ... | | 著者 | Zhang, C, Liu, P, Fan, S. | | 登録日 | 2023-08-29 | | 公開日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | SMPDL3A is a cGAMP-degrading enzyme induced by LXR-mediated lipid metabolism to restrict cGAS-STING DNA sensing.
Immunity, 56, 2023
|
|
3SWD
 
 | | E. coli MurA in complex with UDP-N-acetylmuramic acid and covalent adduct of PEP with Cys115 | | 分子名称: | (2R)-2-{[(2R,3R,4R,5S,6R)-3-(acetylamino)-2-{[(S)-{[(R)-{[(2R,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}-5-hydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-4-yl]oxy}propanoic acid, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | 著者 | Zhu, J.-Y, Schonbrunn, E. | | 登録日 | 2011-07-13 | | 公開日 | 2012-03-14 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Functional Consequence of Covalent Reaction of Phosphoenolpyruvate with UDP-N-acetylglucosamine 1-Carboxyvinyltransferase (MurA).
J.Biol.Chem., 287, 2012
|
|
6DJC
 
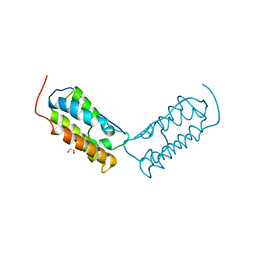 | | Crystal structure of human Bromodomain-containing protein 4 (BRD4) bromodomain with MS645 | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N,N'-(decane-1,10-diyl)bis{2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetamide} | | 著者 | Ren, C, Zhou, M.M. | | 登録日 | 2018-05-25 | | 公開日 | 2018-07-25 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8YLC
 
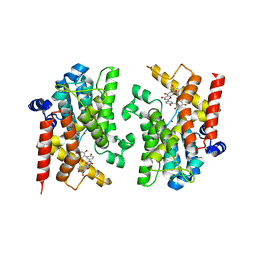 | | The crystal structure of PDE4D with Amentoflavone | | 分子名称: | 3',5'-cyclic-AMP phosphodiesterase 4D, 8-[5-[5,7-bis(oxidanyl)-4-oxidanylidene-chromen-2-yl]-2-oxidanyl-phenyl]-2-(4-hydroxyphenyl)-5,7-bis(oxidanyl)chromen-4-one, MAGNESIUM ION, ... | | 著者 | Huang, Y.-Y, Luo, H.-B. | | 登録日 | 2024-03-06 | | 公開日 | 2024-06-12 | | 実験手法 | X-RAY DIFFRACTION (2.30003715 Å) | | 主引用文献 | Discovery of amentoflavone as a natural PDE4 inhibitor with anti-fibrotic effects
Chin.Chem.Lett., 2024
|
|
6T2K
 
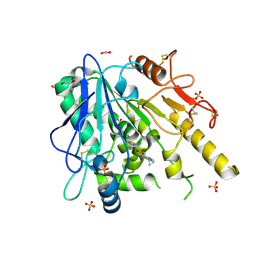 | | Furano[2,3-d]prymidine amides as Notum inhibitors | | 分子名称: | 1,2-ETHANEDIOL, 2-(6-chloranyl-7-cyclopropyl-thieno[3,2-d]pyrimidin-4-yl)sulfanylethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhao, Y, Jones, E.Y. | | 登録日 | 2019-10-08 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Scaffold-hopping identifies furano[2,3-d]pyrimidine amides as potent Notum inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
7M6F
 
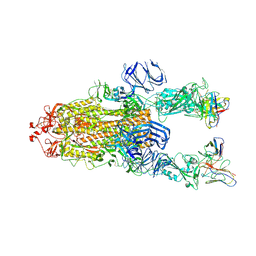 | |
7M6G
 
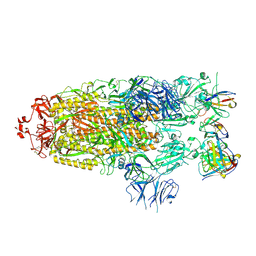 | |
7M6I
 
 | |
7M6E
 
 | |
7M6H
 
 | |
7M6D
 
 | |
8WH8
 
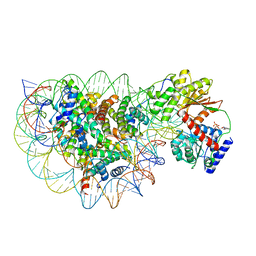 | | Structure of DDM1-nucleosome complex in ADP state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, DNA (antisense strand), ... | | 著者 | Liu, Y, Zhang, Z, Du, J. | | 登録日 | 2023-09-22 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WHB
 
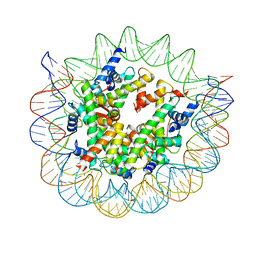 | | Structure of nucleosome core particle of Arabidopsis thaliana | | 分子名称: | DNA (antisense strand), DNA (sense strand), Histone H2A.6, ... | | 著者 | Liu, Y, Zhang, Z, Du, J. | | 登録日 | 2023-09-23 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.17 Å) | | 主引用文献 | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
5H7U
 
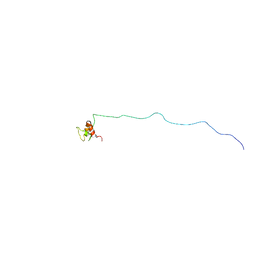 | | NMR structure of eIF3 36-163 | | 分子名称: | Eukaryotic translation initiation factor 3 subunit C | | 著者 | Nagata, T, Obayashi, E. | | 登録日 | 2016-11-21 | | 公開日 | 2017-05-31 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Molecular Landscape of the Ribosome Pre-initiation Complex during mRNA Scanning: Structural Role for eIF3c and Its Control by eIF5.
Cell Rep, 18, 2017
|
|
6XFP
 
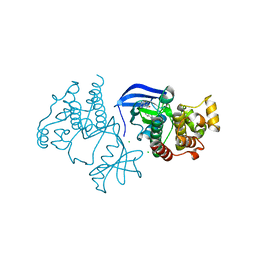 | | Crystal Structure of BRAF kinase domain bound to Belvarafenib | | 分子名称: | 4-amino-N-{1-[(3-chloro-2-fluorophenyl)amino]-6-methylisoquinolin-5-yl}thieno[3,2-d]pyrimidine-7-carboxamide, CHLORIDE ION, Serine/threonine-protein kinase B-raf | | 著者 | Yin, J, Sudhamsu, J. | | 登録日 | 2020-06-16 | | 公開日 | 2021-03-10 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | ARAF mutations confer resistance to the RAF inhibitor belvarafenib in melanoma.
Nature, 594, 2021
|
|
8YGJ
 
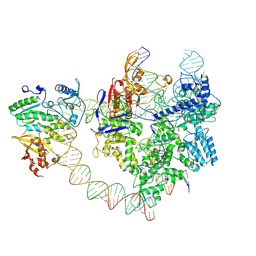 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 28-nt) | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(P*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | 著者 | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | 登録日 | 2024-02-26 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUU
 
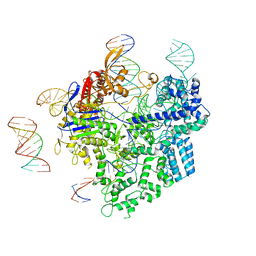 | | SpCas9-pegRNA-target DNA complex (pre-initiation) | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (34-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | 著者 | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | 登録日 | 2023-10-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUS
 
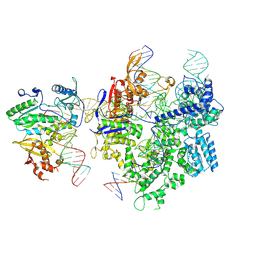 | | SpCas9-MMLV RT-pegRNA-target DNA complex (termination) | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (40-MER), DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), ... | | 著者 | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | 登録日 | 2023-10-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.9 Å) | | 主引用文献 | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUV
 
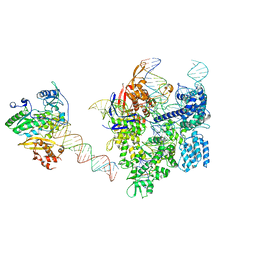 | | SpCas9-MMLV RT-pegRNA-target DNA complex (elongation 16-nt) | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (50-MER), ... | | 著者 | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | 登録日 | 2023-10-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WUT
 
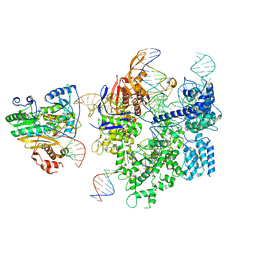 | | SpCas9-MMLV RT-pegRNA-target DNA complex (initiation) | | 分子名称: | CRISPR-associated endonuclease Cas9/Csn1, DNA (5'-D(*TP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*TP*AP*CP*TP*AP*G)-3'), DNA (51-MER), ... | | 著者 | Shuto, Y, Nakagawa, R, Hoki, M, Omura, S.N, Hirano, H, Itoh, Y, Nureki, O. | | 登録日 | 2023-10-21 | | 公開日 | 2024-06-05 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Structural basis for pegRNA-guided reverse transcription by a prime editor.
Nature, 631, 2024
|
|
8WH5
 
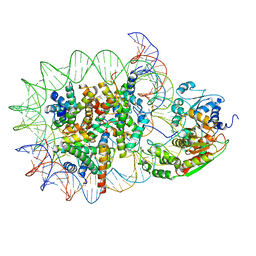 | | Structure of DDM1-nucleosome complex in the apo state | | 分子名称: | ATP-dependent DNA helicase DDM1, DNA (antisense strand), DNA (sense strand), ... | | 著者 | Liu, Y, Zhang, Z, Du, J. | | 登録日 | 2023-09-22 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.58 Å) | | 主引用文献 | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WHA
 
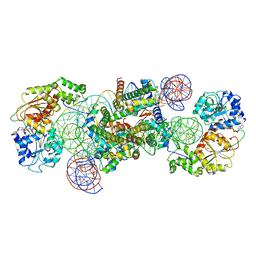 | | Structure of DDM1-nucleosome complex in the ADP-BeFx state with DDM1 bound to SHL2 and SHL-2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | 著者 | Liu, Y, Zhang, Z, Du, J. | | 登録日 | 2023-09-22 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
8WH9
 
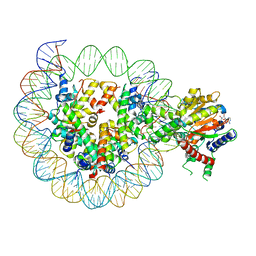 | | Structure of DDM1-nucleosome complex in ADP-BeFx state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent DNA helicase DDM1, BERYLLIUM TRIFLUORIDE ION, ... | | 著者 | Liu, Y, Zhang, Z, Du, J. | | 登録日 | 2023-09-22 | | 公開日 | 2024-04-17 | | 実験手法 | ELECTRON MICROSCOPY (3.31 Å) | | 主引用文献 | Molecular basis of chromatin remodelling by DDM1 involved in plant DNA methylation.
Nat.Plants, 10, 2024
|
|
