6UCE
 
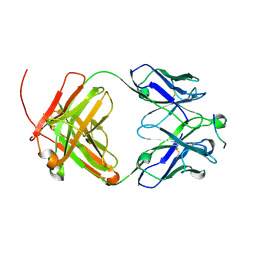 | |
7LQV
 
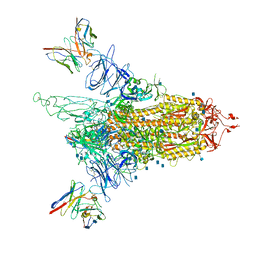 | | Cryo-EM structure of NTD-directed neutralizing antibody 4-8 Fab in complex with SARS-CoV-2 S2P spike | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-8 Heavy Chain, 4-8 Light chain, ... | | 著者 | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | 登録日 | 2021-02-15 | | 公開日 | 2021-03-24 | | 実験手法 | ELECTRON MICROSCOPY (3.25 Å) | | 主引用文献 | Potent SARS-CoV-2 Neutralizing Antibodies Directed Against Spike N-Terminal Domain Target a Single Supersite
Cell Host Microbe, 2021
|
|
7LQW
 
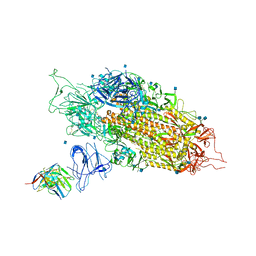 | | Cryo-EM structure of NTD-directed neutralizing antibody 2-17 Fab in complex with SARS-CoV-2 S2P spike | | 分子名称: | 2-17 Heavy Chain, 2-17 Light Chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | 登録日 | 2021-02-15 | | 公開日 | 2021-03-24 | | 最終更新日 | 2021-05-26 | | 実験手法 | ELECTRON MICROSCOPY (4.47 Å) | | 主引用文献 | Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite.
Cell Host Microbe, 29, 2021
|
|
6UCF
 
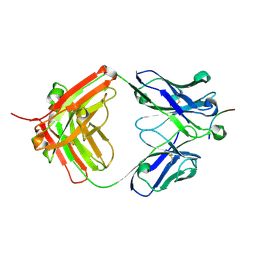 | |
6VIL
 
 | |
4WD7
 
 | |
8IGO
 
 | | Crystal structure of apo SARS-CoV-2 main protease | | 分子名称: | 3C-like proteinase nsp5 | | 著者 | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | 登録日 | 2023-02-21 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
8IGN
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with RAY1216 | | 分子名称: | (3~{S},3~{a}~{S},6~{a}~{R})-2-[(2~{S})-2-cyclohexyl-2-[2,2,2-tris(fluoranyl)ethanoylamino]ethanoyl]-~{N}-[(2~{S})-4-(cyclopentylamino)-3,4-bis(oxidanylidene)-1-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]butan-2-yl]-3,3~{a},4,5,6,6~{a}-hexahydro-1~{H}-cyclopenta[c]pyrrole-3-carboxamide, 3C-like proteinase nsp5 | | 著者 | Huang, X, Zhou, B, Xu, J, Yang, Z, Zhong, N, Xiong, X. | | 登録日 | 2023-02-21 | | 公開日 | 2023-04-05 | | 最終更新日 | 2024-04-17 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Preclinical evaluation of the SARS-CoV-2 M pro inhibitor RAY1216 shows improved pharmacokinetics compared with nirmatrelvir.
Nat Microbiol, 9, 2024
|
|
7L5B
 
 | |
6WXK
 
 | | PHF23 PHD Domain Apo | | 分子名称: | PHD finger protein 23, ZINC ION | | 著者 | Vann, K.R, Zhang, J, Zhang, Y, Kutateladze, T. | | 登録日 | 2020-05-11 | | 公開日 | 2020-07-15 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Mechanistic insights into chromatin targeting by leukemic NUP98-PHF23 fusion.
Nat Commun, 11, 2020
|
|
6U8X
 
 | | Crystal structure of DNMT3B-DNMT3L in complex with CpApG DNA | | 分子名称: | CpApG DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | 著者 | Gao, L, Song, J. | | 登録日 | 2019-09-06 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.95063877 Å) | | 主引用文献 | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
6U8P
 
 | | Crystal structure of DNMT3B-DNMT3L in complex with CpGpA DNA | | 分子名称: | CpGpA DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | 著者 | Gao, L, Song, J. | | 登録日 | 2019-09-05 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.05 Å) | | 主引用文献 | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
6U8V
 
 | | Crystal structure of DNMT3B-DNMT3L in complex with CpGpT DNA | | 分子名称: | CpGpT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | 著者 | Gao, L, Zhang, Z.M, Song, J. | | 登録日 | 2019-09-06 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
6U8W
 
 | | Crystal structure of DNMT3B(K777A)-DNMT3L in complex with CpGpT DNA | | 分子名称: | CpGpT DNA (25-MER), DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3B, ... | | 著者 | Gao, L, Zhang, Z.M, Song, J. | | 登録日 | 2019-09-06 | | 公開日 | 2020-06-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.94891548 Å) | | 主引用文献 | Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms.
Nat Commun, 11, 2020
|
|
2ICW
 
 | | Crystal structure of a complete ternary complex between TCR, superantigen, and peptide-MHC class II molecule | | 分子名称: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-1 beta chain, ... | | 著者 | Wang, L, Zhao, Y, Li, H. | | 登録日 | 2006-09-13 | | 公開日 | 2007-03-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Crystal structure of a complete ternary complex of TCR, superantigen and peptide-MHC.
Nat.Struct.Mol.Biol., 14, 2007
|
|
5WUF
 
 | | Structural basis for conductance through TRIC cation channels | | 分子名称: | CADMIUM ION, Putative membrane protein | | 著者 | Mao, Y, Gao, F, Su, M, Wang, X.H, Zeng, Y, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | 登録日 | 2016-12-17 | | 公開日 | 2017-08-09 | | 実験手法 | X-RAY DIFFRACTION (2.401 Å) | | 主引用文献 | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
3PMT
 
 | | Crystal structure of the Tudor domain of human Tudor domain-containing protein 3 | | 分子名称: | TETRAETHYLENE GLYCOL, Tudor domain-containing protein 3 | | 著者 | Lam, R, Bian, C.B, Guo, Y.H, Xu, C, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2010-11-18 | | 公開日 | 2010-12-01 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of TDRD3 and Methyl-Arginine Binding Characterization of TDRD3, SMN and SPF30.
Plos One, 7, 2012
|
|
7JM5
 
 | | Crystal structure of KDM4B in complex with QC6352 | | 分子名称: | 3-[({(1R)-6-[methyl(phenyl)amino]-1,2,3,4-tetrahydronaphthalen-1-yl}methyl)amino]pyridine-4-carboxylic acid, Lysine-specific demethylase 4B, NICKEL (II) ION, ... | | 著者 | White, S.W, Yun, M. | | 登録日 | 2020-07-31 | | 公開日 | 2022-02-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Targeting KDM4 for treating PAX3-FOXO1-driven alveolar rhabdomyosarcoma.
Sci Transl Med, 14, 2022
|
|
4DJG
 
 | |
5WCH
 
 | | Crystal structure of the catalytic domain of human USP9X | | 分子名称: | Probable ubiquitin carboxyl-terminal hydrolase FAF-X, UNKNOWN ATOM OR ION, ZINC ION | | 著者 | Dong, A, Zhang, Q, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | 登録日 | 2017-06-30 | | 公開日 | 2018-07-04 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure and activity-based labeling reveal the mechanisms for linkage-specific substrate recognition by deubiquitinase USP9X.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4DIX
 
 | |
6L42
 
 | |
8G59
 
 | | Cryo-EM structure of the TUG891 bound GPR120-Giq complex | | 分子名称: | 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | 著者 | Mao, C, Xiao, P, Tao, X, Qin, J, He, Q, Zhang, C, Yu, X, Zhang, Y, Sun, J. | | 登録日 | 2023-02-12 | | 公開日 | 2023-03-08 | | 最終更新日 | 2023-04-19 | | 実験手法 | ELECTRON MICROSCOPY (2.64 Å) | | 主引用文献 | Unsaturated bond recognition leads to biased signal in a fatty acid receptor.
Science, 380, 2023
|
|
4WD8
 
 | |
8DS8
 
 | |
