8SLN
 
 | |
6YSK
 
 | | 1-phenylpyrroles and 1-enylpyrrolidines as inhibitors of Notum | | 分子名称: | (3~{S})-1-[4-chloranyl-3-(trifluoromethyl)phenyl]pyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhao, Y, Jones, E.Y, Fish, P. | | 登録日 | 2020-04-22 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.21 Å) | | 主引用文献 | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
8SLM
 
 | | Crystal structure of Deinococcus geothermalis PprI | | 分子名称: | MANGANESE (II) ION, SULFATE ION, Zn dependent hydrolase fused to HTH domain, ... | | 著者 | Zhao, Y, Lu, H. | | 登録日 | 2023-04-23 | | 公開日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.81 Å) | | 主引用文献 | The Deinococcus protease PprI senses DNA damage by directly interacting with single-stranded DNA.
Nat Commun, 15, 2024
|
|
4USG
 
 | | Crystal structure of PC4 W89Y mutant complex with DNA | | 分子名称: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP *TP*TP*TP*TP*TP*G)-3', ACTIVATED RNA POLYMERASE II TRANSCRIPTIONAL COACTIVATOR P15 | | 著者 | Zhao, Y, Liu, J. | | 登録日 | 2014-07-08 | | 公開日 | 2015-03-18 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.973 Å) | | 主引用文献 | Substitution of Tryptophan 89 with Tyrosine Switches the DNA Binding Mode of Pc4.
Sci.Rep., 5, 2015
|
|
8S7C
 
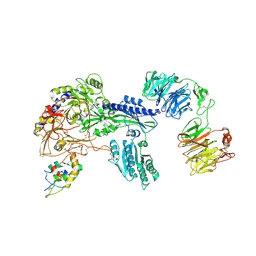 | | Ternary Complex of Cachd1, FZD5 and LRP6 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Frizzled-5, ... | | 著者 | Zhao, Y, Ren, J, Jones, E.Y. | | 登録日 | 2024-02-29 | | 公開日 | 2024-04-24 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (4.7 Å) | | 主引用文献 | Cachd1 interacts with Wnt receptors and regulates neuronal asymmetry in the zebrafish brain.
Science, 384, 2024
|
|
6DB9
 
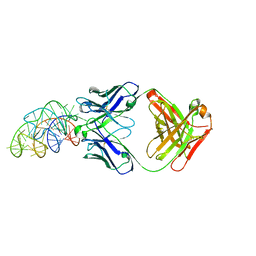 | | Structural basis for promiscuous binding and activation of fluorogenic dyes by DIR2s RNA aptamer | | 分子名称: | Fab-Heavy-Chain, Fab-Light-Chain, MAGNESIUM ION, ... | | 著者 | Shao, Y, Shelke, S.A, Laski, A, Piccirilli, J.A. | | 登録日 | 2018-05-02 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.025 Å) | | 主引用文献 | Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif.
Nat Commun, 9, 2018
|
|
6DB8
 
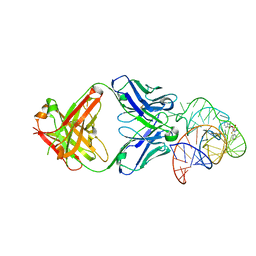 | | Structural basis for promiscuous binding and activation of fluorogenic dyes by DIR2s RNA aptamer | | 分子名称: | 2-[(Z)-(3-methyl-1,3-benzoxazol-2(3H)-ylidene)methyl]-3-(3-sulfopropyl)-1,3-benzothiazol-3-ium, Fab-Heavy chain, Fab-Light chain, ... | | 著者 | Shao, Y, Shelke, S.A, Laski, A, Piccirilli, J.A. | | 登録日 | 2018-05-02 | | 公開日 | 2018-11-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.86541343 Å) | | 主引用文献 | Structural basis for activation of fluorogenic dyes by an RNA aptamer lacking a G-quadruplex motif.
Nat Commun, 9, 2018
|
|
6M03
 
 | | The crystal structure of COVID-19 main protease in apo form | | 分子名称: | 3C-like proteinase | | 著者 | Zhang, B, Zhao, Y, Jin, Z, Liu, X, Yang, H, Rao, Z. | | 登録日 | 2020-02-19 | | 公開日 | 2020-03-11 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6R8P
 
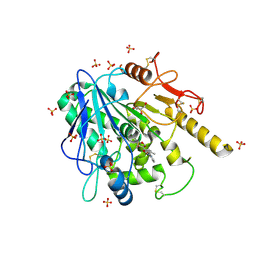 | | Notum fragment 723 | | 分子名称: | 2-(2-methylphenoxy)-~{N}-pyridin-3-yl-ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | 著者 | Zhao, Y, Jones, E.Y. | | 登録日 | 2019-04-02 | | 公開日 | 2019-05-08 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Discovery of 2-phenoxyacetamides as inhibitors of the Wnt-depalmitoleating enzyme NOTUM from an X-ray fragment screen.
Medchemcomm, 10, 2019
|
|
6T2H
 
 | | Furano[2,3-d]prymidine amides as Notum inhibitors | | 分子名称: | 1,2-ETHANEDIOL, 2-[[(4~{S})-5-chloranyl-6-methyl-1,2,3,4-tetrahydrothieno[2,3-d]pyrimidin-4-yl]sulfanyl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhao, Y, Jones, E.Y. | | 登録日 | 2019-10-08 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.41 Å) | | 主引用文献 | Scaffold-hopping identifies furano[2,3-d]pyrimidine amides as potent Notum inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6T2K
 
 | | Furano[2,3-d]prymidine amides as Notum inhibitors | | 分子名称: | 1,2-ETHANEDIOL, 2-(6-chloranyl-7-cyclopropyl-thieno[3,2-d]pyrimidin-4-yl)sulfanylethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhao, Y, Jones, E.Y. | | 登録日 | 2019-10-08 | | 公開日 | 2020-01-01 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.38 Å) | | 主引用文献 | Scaffold-hopping identifies furano[2,3-d]pyrimidine amides as potent Notum inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
4XWW
 
 | | Crystal structure of RNase J complexed with RNA | | 分子名称: | DR2417, GLYCEROL, MANGANESE (II) ION, ... | | 著者 | Lu, M, Zhang, H, Xu, Q, Hua, Y, Zhao, Y. | | 登録日 | 2015-01-29 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J
Nucleic Acids Res., 43, 2015
|
|
4XWT
 
 | | Crystal structure of RNase J complexed with UMP | | 分子名称: | DR2417, GLYCEROL, MANGANESE (II) ION, ... | | 著者 | Lu, M, Zhang, H, Xu, Q, Hua, Y, Zhao, Y. | | 登録日 | 2015-01-29 | | 公開日 | 2015-12-16 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.003 Å) | | 主引用文献 | Structural insights into catalysis and dimerization enhanced exonuclease activity of RNase J
Nucleic Acids Res., 43, 2015
|
|
7SUK
 
 | | Structure of Bfr2-Lcp5 Complex Observed in the Small Subunit Processome Isolated from R2TP-depleted Yeast Cells | | 分子名称: | 18S pre-rRNA, 40S ribosomal protein S11-A, 40S ribosomal protein S13, ... | | 著者 | Rai, J, Zhao, Y, Li, H. | | 登録日 | 2021-11-17 | | 公開日 | 2022-07-06 | | 最終更新日 | 2023-08-16 | | 実験手法 | ELECTRON MICROSCOPY (3.99 Å) | | 主引用文献 | Artificial intelligence-assisted cryoEM structure of Bfr2-Lcp5 complex observed in the yeast small subunit processome.
Commun Biol, 5, 2022
|
|
7BVH
 
 | | Crystal structure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose | | 分子名称: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, Meromycolate extension acyl carrier protein, ... | | 著者 | Zhao, Y, Zhang, L, Wu, L.J, Wang, Q, Li, J, Besra, G.S, Rao, Z.H. | | 登録日 | 2020-04-10 | | 公開日 | 2020-04-29 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (3.3 Å) | | 主引用文献 | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7VAH
 
 | |
7BUY
 
 | | The crystal structure of COVID-19 main protease in complex with carmofur | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, hexylcarbamic acid | | 著者 | Zhao, Y, Zhang, B, Jin, Z, Liu, X, Yang, H, Rao, Z. | | 登録日 | 2020-04-08 | | 公開日 | 2020-04-29 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural basis for the inhibition of SARS-CoV-2 main protease by antineoplastic drug carmofur.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7MPI
 
 | | Stm1 bound vacant 80S structure isolated from cbf5-D95A | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | 著者 | Rai, J, Zhao, Y, Li, H. | | 登録日 | 2021-05-04 | | 公開日 | 2022-05-11 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
7MPJ
 
 | | Stm1 bound vacant 80S structure isolated from wild-type | | 分子名称: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | 著者 | Rai, J, Zhao, Y, Li, H. | | 登録日 | 2021-05-04 | | 公開日 | 2022-05-11 | | 最終更新日 | 2023-12-13 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
7E5X
 
 | |
7N8B
 
 | | Cycloheximide bound vacant 80S structure isolated from cbf5-D95A | | 分子名称: | 18S RIBOSOMAL RNA, 25S, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | 著者 | Rai, J, Zhao, Y, Li, H. | | 登録日 | 2021-06-14 | | 公開日 | 2022-05-11 | | 最終更新日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.05 Å) | | 主引用文献 | CryoEM structures of pseudouridine-free ribosome suggest impacts of chemical modifications on ribosome conformations.
Structure, 30, 2022
|
|
7DVP
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp4|5 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp4/5 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-14 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW0
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp14|15 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp14/15 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DW6
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp15|16 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp15/16 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7DVY
 
 | | SARS-CoV-2 Mpro mutant (H41A) in complex with nsp9|10 peptidyl substrate | | 分子名称: | 3C-like proteinase, nsp9/10 peptidyl substrate | | 著者 | Liu, X, Zhao, Y, Yang, H, Rao, Z. | | 登録日 | 2021-01-15 | | 公開日 | 2022-01-19 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural basis for replicase polyprotein cleavage and substrate specificity of main protease from SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
