1MZK
 
 | | NMR structure of kinase-interacting FHA domain of kinase associated protein phosphatase, KAPP in Arabidopsis | | Descriptor: | KINASE ASSOCIATED PROTEIN PHOSPHATASE | | Authors: | Lee, G, Ding, Z, Walker, J.C, Van Doren, S.R. | | Deposit date: | 2002-10-08 | | Release date: | 2003-09-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the forkhead-associated domain from the Arabidopsis receptor kinase-associated protein phosphatase.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2AX1
 
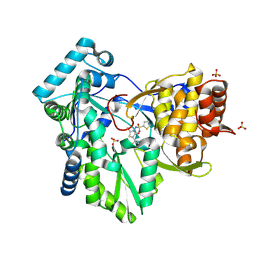 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5ee) | | Descriptor: | 5R-(3,4-DICHLOROPHENYLMETHYL)-3-(2-THIOPHENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | Authors: | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
2AX0
 
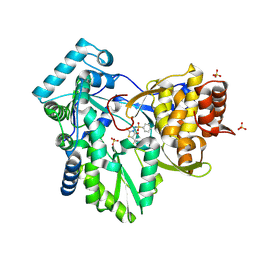 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5x) | | Descriptor: | 5R-(2E-METHYL-3-PHENYL-ALLYL)-3-(BENZENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | Authors: | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
2AWZ
 
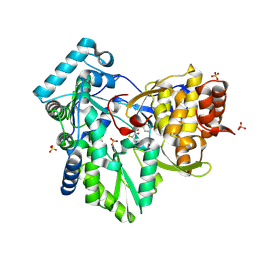 | | Hepatitis C Virus NS5b RNA Polymerase in complex with a covalent inhibitor (5h) | | Descriptor: | 5R-(4-BROMOPHENYLMETHYL)-3-(BENZENESULFONYLAMINO)-4-OXO-2-THIONOTHIAZOLIDINE, Genome polyprotein, SULFATE ION | | Authors: | Powers, J.P, Piper, D.E, Li, Y, Mayorga, V, Anzola, J, Chen, J.M, Jaen, J.C, Lee, G, Liu, J, Peterson, M.G, Tonn, G.R, Ye, Q, Walker, N.P, Wang, Z. | | Deposit date: | 2005-09-02 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | SAR and Mode of Action of Novel Non-Nucleoside Inhibitors of Hepatitis C NS5b RNA Polymerase.
J.Med.Chem., 49, 2006
|
|
7KC3
 
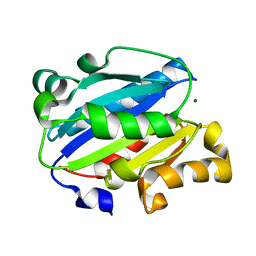 | | X-ray structure of Lfa-1 I domain collected at 273 K | | Descriptor: | Integrin alpha-L, MAGNESIUM ION | | Authors: | Woldeyes, R.A, Hallenbeck, K.K, Pfaff, S.J, Lee, G, Cortez, S.V, Kelly, M.J, Akassoglou, K, Arkin, M.R, Fraser, J.S. | | Deposit date: | 2020-10-05 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Divergent conformational dynamics controls allosteric ligand accessibility across evolutionarily related I-domain-containing integrins
To Be Published
|
|
7KC5
 
 | | X-ray structure of Lfa-1 I domain in complex with BMS-68852 collected at 273 K | | Descriptor: | 6-[(5S,9R)-9-(4-cyanophenyl)-3-(3,5-dichlorophenyl)-1-methyl-2,4-dioxo-1,3,7-triazaspiro[4.4]non-7-yl]pyridine-3-carboxylic acid, Integrin alpha-L, MAGNESIUM ION | | Authors: | Woldeyes, R.A, Hallenbeck, K.K, Pfaff, S.J, Lee, G, Cortez, S.V, Kelly, M.J, Akassoglou, K, Arkin, M.R, Fraser, J.S. | | Deposit date: | 2020-10-05 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Divergent conformational dynamics controls allosteric ligand accessibility across evolutionarily related I-domain-containing integrins
To Be Published
|
|
7KC6
 
 | | X-ray structure of Lfa-1 I domain in complex with Lovastatin collected at 273 K | | Descriptor: | Integrin alpha-L, LOVASTATIN, MAGNESIUM ION | | Authors: | Woldeyes, R.A, Hallenbeck, K.K, Pfaff, S.J, Lee, G, Cortez, S.V, Kelly, M.J, Akassoglou, K, Arkin, M.R, Fraser, J.S. | | Deposit date: | 2020-10-05 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Divergent conformational dynamics controls allosteric ligand accessibility across evolutionarily related I-domain-containing integrins
To Be Published
|
|
7VZM
 
 | | Anti-CRISPR AcrIE4-F7 | | Descriptor: | AcrIE4-F7 | | Authors: | Hong, S.H, Lee, G, Bae, E, Suh, J.Y. | | Deposit date: | 2021-11-16 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of AcrIE4-F7 reveals a common strategy for dual CRISPR inhibition by targeting PAM recognition sites.
Nucleic Acids Res., 50, 2022
|
|
1H1B
 
 | | Crystal structure of human neutrophil elastase complexed with an inhibitor (GW475151) | | Descriptor: | (2S)-3-METHYL-2-((2R,3S)-3-[(METHYLSULFONYL)AMINO]-1-{[2-(PYRROLIDIN-1-YLMETHYL)-1,3-OXAZOL-4-YL]CARBONYL}PYRROLIDIN-2-YL)BUTANOIC ACID, LEUKOCYTE ELASTASE, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Macdonald, S.J.F, Dowle, M.D, Harrison, L.A, Clarke, G.D.E, Inglis, G.G.A, Johnson, M.R, Smith, R.A, Amour, A, Fleetwood, G, Humphreys, D.C, Molloy, C.R, Dixon, M, Godward, R.E, Wonacott, A.J, Singh, O.M.P, Hodgson, S.T, Hardy, G.W. | | Deposit date: | 2002-07-05 | | Release date: | 2002-08-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Further Pyrrolidine Trans-Lactams as Inhibitors of Human Neutrophil Elastase (Hne) with Potential as Development Candidates and the Crystal Structure of Hne Complexed with an Inhibitor (Gw475151)
J.Med.Chem., 45, 2002
|
|
4E4K
 
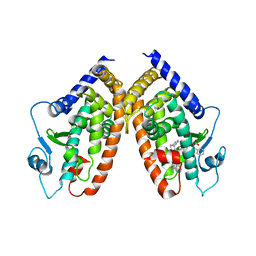 | | Crystal Structure of PPARgamma with the ligand JO21 | | Descriptor: | (2S)-3-phenyl-2-{[2'-(propan-2-yl)biphenyl-4-yl]oxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Loiodice, F, Fracchiolla, G, Laghezza, A, Carbonara, G, Piemontese, L, Lavecchia, A, Novellino, E. | | Deposit date: | 2012-03-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New 2-(Aryloxy)-3-phenylpropanoic Acids as Peroxisome Proliferator-Activated Receptor alpha/gamma Dual Agonists Able To Upregulate Mitochondrial Carnitine Shuttle System Gene Expression.
J.Med.Chem., 56, 2013
|
|
4E4Q
 
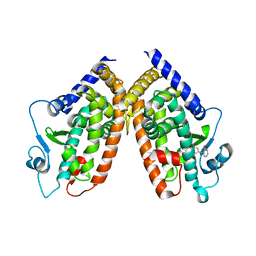 | | Crystal structure of PPARgamma with the ligand FS214 | | Descriptor: | (2R)-3-phenyl-2-{[2'-(propan-2-yl)biphenyl-4-yl]oxy}propanoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Loiodice, F, Fracchiolla, G, Laghezza, A, Carbonara, G, Piemontese, L, Lavecchia, A, Novellino, E. | | Deposit date: | 2012-03-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | New 2-(Aryloxy)-3-phenylpropanoic Acids as Peroxisome Proliferator-Activated Receptor alpha/gamma Dual Agonists Able To Upregulate Mitochondrial Carnitine Shuttle System Gene Expression.
J.Med.Chem., 56, 2013
|
|
3K8S
 
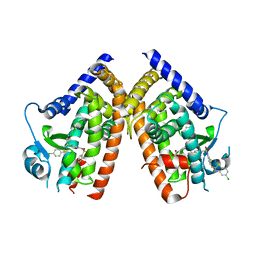 | | Crystal Structure of PPARg in complex with T2384 | | Descriptor: | 2-chloro-N-{3-chloro-4-[(5-chloro-1,3-benzothiazol-2-yl)sulfanyl]phenyl}-4-(trifluoromethyl)benzenesulfonamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Wang, Z. | | Deposit date: | 2009-10-14 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | T2384, a novel antidiabetic agent with unique peroxisome proliferator-activated receptor gamma binding properties
J.Biol.Chem., 283, 2008
|
|
4TT4
 
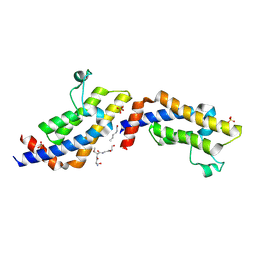 | | Crystal structure of ATAD2A bromodomain complexed with H3(1-21)K14Ac peptide | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, Histone H3(1-21)K4Ac, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TT2
 
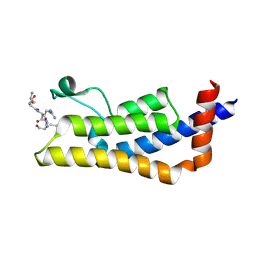 | | Crystal structure of ATAD2A bromodomain complexed with H4(1-20)K5Ac peptide | | Descriptor: | ATPase family AAA domain-containing protein 2, Histone H4K5Ac | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TU6
 
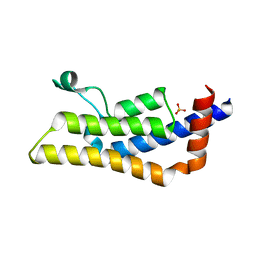 | | Crystal structure of apo ATAD2A bromodomain with N1064 alternate conformation | | Descriptor: | ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-23 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TU4
 
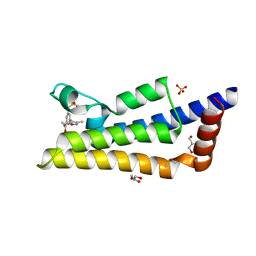 | | Crystal structure of ATAD2A bromodomain complexed with 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(phenylsulfonyl)amino]benzoicacid | | Descriptor: | 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(phenylsulfonyl)amino]benzoic acid, ATPase family AAA domain-containing protein 2, CHLORIDE ION, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-23 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TT6
 
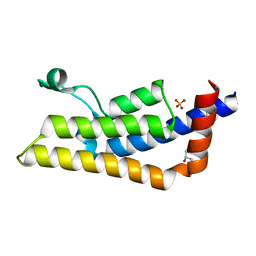 | | Crystal structure of ATAD2A bromodomain double mutant N1063A-Y1064A in apo form | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-19 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
4TTE
 
 | | Crystal structure of ATAD2A bromodomain complexed with methyl 3-amino-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzoate | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-20 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
8VB1
 
 | | Crystal structure of HIV-1 protease with GS-9770 | | Descriptor: | (2S)-2-{(3M)-4-chloro-3-[1-(difluoromethyl)-1H-1,2,4-triazol-5-yl]phenyl}-2-[(2E,4R)-4-[4-(2-cyclopropyl-2H-1,2,3-triazol-4-yl)phenyl]-2-imino-5-oxo-4-(3,3,3-trifluoro-2,2-dimethylpropyl)imidazolidin-1-yl]ethyl [1-(difluoromethyl)cyclopropyl]carbamate, HIV-1 protease | | Authors: | Lansdon, E.B. | | Deposit date: | 2023-12-11 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Preclinical characterization of a non-peptidomimetic HIV protease inhibitor with improved metabolic stability.
Antimicrob.Agents Chemother., 68, 2024
|
|
5Y3N
 
 | | Structure of TRAP1 complexed with DN401 | | Descriptor: | 1-[(6-bromanyl-1,3-benzodioxol-5-yl)methyl]-4-chloranyl-pyrazolo[3,4-d]pyrimidin-6-amine, Heat shock protein 75 kDa, mitochondrial | | Authors: | Jeong, H, Park, H.K, Kang, S, Kang, B.H, Lee, C. | | Deposit date: | 2017-07-29 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Paralog Specificity Determines Subcellular Distribution, Action Mechanism, and Anticancer Activity of TRAP1 Inhibitors.
J. Med. Chem., 60, 2017
|
|
5Y3O
 
 | | Structure of TRAP1 complexed with DN320 | | Descriptor: | 4-chloranyl-1-[(4-methoxy-3,5-dimethyl-pyridin-2-yl)methyl]pyrazolo[3,4-d]pyrimidin-6-amine, Heat shock protein 75 kDa, mitochondrial | | Authors: | Jeong, H, Park, H.K, Kang, S, Kang, B.H, Lee, C. | | Deposit date: | 2017-07-29 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Paralog Specificity Determines Subcellular Distribution, Action Mechanism, and Anticancer Activity of TRAP1 Inhibitors.
J. Med. Chem., 60, 2017
|
|
9B3P
 
 | |
4WAG
 
 | | Phosphatidylinositol 4-kinase III beta crystallized with MI103 inhibitor | | Descriptor: | 6-chloro-3-(3,4-dimethoxyphenyl)-2-methylimidazo[1,2-b]pyridazin-8-amine, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta | | Authors: | Chalupska, D, Boura, E. | | Deposit date: | 2014-08-29 | | Release date: | 2015-05-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.407 Å) | | Cite: | Highly Selective Phosphatidylinositol 4-Kinase III beta Inhibitors and Structural Insight into Their Mode of Action.
J.Med.Chem., 58, 2015
|
|
4WAE
 
 | | Phosphatidylinositol 4-kinase III beta crystallized with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta | | Authors: | Chalupska, D, Boura, E. | | Deposit date: | 2014-08-29 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.318 Å) | | Cite: | Highly Selective Phosphatidylinositol 4-Kinase III beta Inhibitors and Structural Insight into Their Mode of Action.
J.Med.Chem., 58, 2015
|
|
3NQW
 
 | | A metazoan ortholog of SpoT hydrolyzes ppGpp and plays a role in starvation responses | | Descriptor: | CG11900, MANGANESE (II) ION, SULFATE ION | | Authors: | Sun, D.W, Kim, H.Y, Kim, K.J, Jeon, Y.H, Chung, J. | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses
Nat.Struct.Mol.Biol., 17, 2010
|
|
