3K55
 
 | | Structure of beta hairpin deletion mutant of beta toxin from Staphylococcus aureus | | Descriptor: | Beta-hemolysin, CHLORIDE ION, SODIUM ION | | Authors: | Kruse, A.C, Huseby, M, Shi, K, Digre, J, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2009-10-06 | | Release date: | 2011-01-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Structure of a mutant beta toxin from Staphylococcus aureus reveals domain swapping and conformational flexibility
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4MQT
 
 | | Structure of active human M2 muscarinic acetylcholine receptor bound to the agonist iperoxo and allosteric modulator LY2119620 | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Muscarinic acetylcholine receptor M2, ... | | Authors: | Kruse, A.C, Ring, A.M, Manglik, A, Hu, J, Hu, K, Eitel, K, Huebner, H, Pardon, E, Valant, C, Sexton, P.M, Christopoulos, A, Felder, C.C, Gmeiner, P, Steyaert, J, Weis, W.I, Garcia, K.C, Wess, J, Kobilka, B.K. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nature, 504, 2013
|
|
4MQS
 
 | | Structure of active human M2 muscarinic acetylcholine receptor bound to the agonist iperoxo | | Descriptor: | 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Muscarinic acetylcholine receptor M2, Nanobody 9-8 | | Authors: | Kruse, A.C, Ring, A.M, Manglik, A, Hu, J, Hu, K, Eitel, K, Huebner, H, Pardon, E, Valant, C, Sexton, P.M, Christopoulos, A, Felder, C.C, Gmeiner, P, Steyaert, J, Weis, W.I, Garcia, K.C, Wess, J, Kobilka, B.K. | | Deposit date: | 2013-09-16 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nature, 504, 2013
|
|
4DAJ
 
 | | Structure of the M3 Muscarinic Acetylcholine Receptor | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, Muscarinic acetylcholine receptor M3, Lysozyme, ... | | Authors: | Kruse, A.C, Hu, J, Pan, A.C, Arlow, D.H, Rosenbaum, D.M, Rosemond, E, Green, H.F, Liu, T, Chae, P.S, Dror, R.O, Shaw, D.E, Weis, W.I, Wess, J, Kobilka, B. | | Deposit date: | 2012-01-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and dynamics of the M3 muscarinic acetylcholine receptor.
Nature, 482, 2012
|
|
3HL6
 
 | | Staphylococcus aureus pathogenicity island 3 ORF9 protein | | Descriptor: | CHLORIDE ION, Pathogenicity island protein | | Authors: | Kruse, A.C, Huseby, M, Shi, K, Digre, J, Schlievert, P.M, Ohlendorf, D.H, Earhart, C.A. | | Deposit date: | 2009-05-26 | | Release date: | 2010-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional studies of a pathogenicity island protein from Staphylococcus aureus
To be Published
|
|
5VNV
 
 | | Crystal structure of Nb.b201 | | Descriptor: | FORMIC ACID, Nb.b201, PENTAETHYLENE GLYCOL, ... | | Authors: | Kruse, A.C, McMahon, C. | | Deposit date: | 2017-05-01 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Yeast surface display platform for rapid discovery of conformationally selective nanobodies.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3QIW
 
 | | Crystal structure of the 226 TCR in complex with MCC-p5E/I-Ek | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H-2 CLASS II HISTOCOMPATIBILITY ANTIGEN, E-K alpha chain, ... | | Authors: | Kruse, A.C, Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
3QIU
 
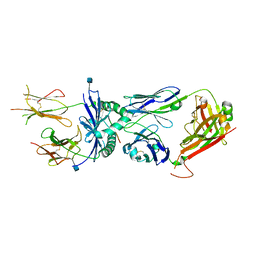 | | Crystal structure of the 226 TCR in complex with MCC/I-Ek | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H-2 CLASS II HISTOCOMPATIBILITY ANTIGEN, E-K alpha chain, ... | | Authors: | Kruse, A.C, Ely, L.K, Newell, E.W, Davis, M.M, Garcia, K.C. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of specificity and cross-reactivity in T cell receptors specific for cytochrome c-I-E(k).
J.Immunol., 186, 2011
|
|
4DKL
 
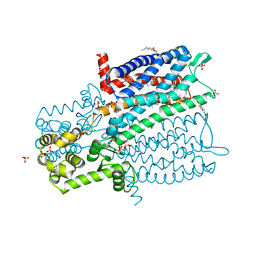 | | Crystal structure of the mu-opioid receptor bound to a morphinan antagonist | | Descriptor: | CHLORIDE ION, CHOLESTEROL, Mu-type opioid receptor, ... | | Authors: | Manglik, A, Kruse, A.C, Kobilka, T.S, Thian, F.S, Mathiesen, J.M, Sunahara, R.K, Pardo, L, Weis, W.I, Kobilka, B.K, Granier, S. | | Deposit date: | 2012-02-03 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the {mu}-opioid receptor bound to a morphinan antagonist.
Nature, 485, 2012
|
|
4QKX
 
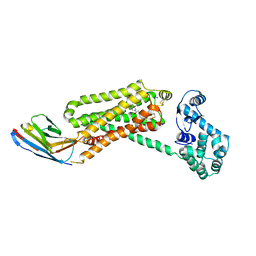 | | Structure of beta2 adrenoceptor bound to a covalent agonist and an engineered nanobody | | Descriptor: | 4-[(1R)-1-hydroxy-2-({2-[3-methoxy-4-(2-sulfanylethoxy)phenyl]ethyl}amino)ethyl]benzene-1,2-diol, Beta-2 adrenergic receptor, R9 protein, ... | | Authors: | Weichert, D, Kruse, A.C, Manglik, A, Hiller, C, Zhang, C, Huebner, H, Kobilka, B.K, Gmeiner, P. | | Deposit date: | 2014-06-10 | | Release date: | 2014-07-23 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Covalent agonists for studying G protein-coupled receptor activation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7JIC
 
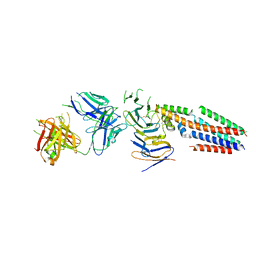 | | Structure of human CD19-CD81 co-receptor complex bound to coltuximab Fab fragment | | Descriptor: | B-lymphocyte antigen CD19, CD81 antigen, Coltuximab Heavy Chain, ... | | Authors: | Susa, K.J, Rawson, S, Kruse, A.C, Blacklow, S.C. | | Deposit date: | 2020-07-23 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structure of the B cell co-receptor CD19 bound to the tetraspanin CD81
Science, 371, 2021
|
|
8UFH
 
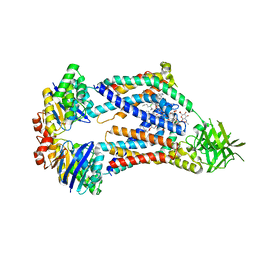 | | Acinetobacter baylyi LptB2FG bound to Acinetobacter baylyi lipopolysaccharide and a macrocyclic peptide | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-2-[(2~{R},4~{R},5~{R},6~{R})-5-[(2~{S},4~{R},5~{R},6~{R})-4-[(2~{R},3~{R},4~{R},5~{S},6~{S})-3-acetamido-6-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-5-oxidanyl-oxan-2-yl]oxy-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-4-[(3~{S})-3-dodecanoyloxydodecanoyl]oxy-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-heptanoyloxynonanoyl]amino]-3-oxidanyl-4-[(3~{R})-3-oxidanyloctanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-5-[[(3~{S})-3-[(3~{R})-3-oxidanyldodecanoyl]oxydecanoyl]amino]-3-phosphonooxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4,5-bis(oxidanyl)oxane-2-carboxylic acid, (7S,10S,13S,17P)-10-(4-aminobutyl)-7-(3-aminopropyl)-17-(6-aminopyridin-3-yl)-20-chloro-13-[(1H-indol-3-yl)methyl]-12-methyl-6,7,9,10,12,13,15,16-octahydropyrido[2,3-b][1,5,8,11,14]benzothiatetraazacycloheptadecine-8,11,14(5H)-trione, LPS export ABC transporter permease LptG, ... | | Authors: | Pahil, K.S, Gilman, M.S.A, Baidin, V, Kruse, A.C, Kahne, D. | | Deposit date: | 2023-10-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A new antibiotic traps lipopolysaccharide in its intermembrane transporter.
Nature, 625, 2024
|
|
8UFG
 
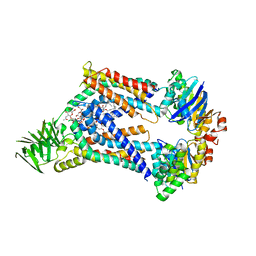 | | Acinetobacter baylyi LptB2FG bound to Acinetobacter baylyi lipopolysaccharide | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-2-[(2~{R},4~{R},5~{R},6~{R})-5-[(2~{S},4~{R},5~{R},6~{R})-4-[(2~{R},3~{R},4~{R},5~{S},6~{S})-3-acetamido-6-carboxy-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-5-oxidanyl-oxan-2-yl]oxy-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-4-[(3~{S})-3-dodecanoyloxydodecanoyl]oxy-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-heptanoyloxyundecanoyl]amino]-3-oxidanyl-4-[(3~{R})-3-oxidanyloctanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-5-[[(3~{S})-3-[(3~{R})-3-oxidanyldecanoyl]oxydecanoyl]amino]-3-phosphonooxy-oxan-2-yl]methoxy]oxan-4-yl]oxy-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4,5-bis(oxidanyl)oxane-2-carboxylic acid, DODECYL-BETA-D-MALTOSIDE, LPS export ABC transporter permease LptG, ... | | Authors: | Pahil, K.S, Gilman, M.S.A, Baidin, V, Kruse, A.C, Kahne, D. | | Deposit date: | 2023-10-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A new antibiotic traps lipopolysaccharide in its intermembrane transporter.
Nature, 625, 2024
|
|
5VNW
 
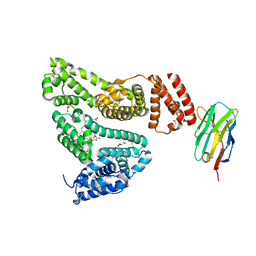 | |
8TH4
 
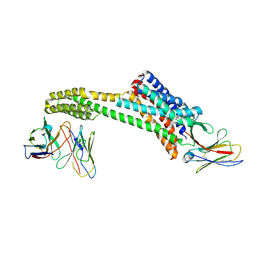 | |
8TH3
 
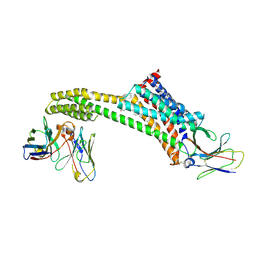 | |
3JS1
 
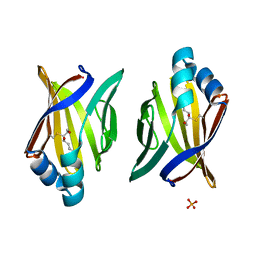 | | Crystal structure of adipocyte fatty acid binding protein covalently modified with 4-hydroxy-2-nonenal | | Descriptor: | Adipocyte fatty acid-binding protein, PHOSPHATE ION | | Authors: | Hellberg, K, Grimsrud, P.A, Kruse, A.C, Banaszak, L.J, Ohlendorf, D.H, Bernlohr, D.A. | | Deposit date: | 2009-09-09 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | X-ray crystallographic analysis of adipocyte fatty acid binding protein (aP2) modified with 4-hydroxy-2-nonenal.
Protein Sci., 19, 2010
|
|
3JSQ
 
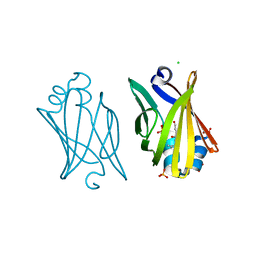 | | Crystal structure of adipocyte fatty acid binding protein non-covalently modified with 4-hydroxy-2-nonenal | | Descriptor: | (2E,4R)-4-HYDROXYNON-2-ENAL, Adipocyte fatty acid-binding protein, CHLORIDE ION, ... | | Authors: | Hellberg, K, Grimsrud, P.A, Kruse, A.C, Banaszak, L.J, Ohlendorf, D.H, Bernlohr, D.A. | | Deposit date: | 2009-09-10 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallographic analysis of adipocyte fatty acid binding protein (aP2) modified with 4-hydroxy-2-nonenal.
Protein Sci., 19, 2010
|
|
6OS1
 
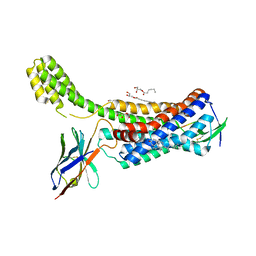 | | Structure of synthetic nanobody-stabilized angiotensin II type 1 receptor bound to TRV023 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, Nanobody Nb.AT110i1_le, ... | | Authors: | Wingler, L.M, Staus, D.P, Skiba, M.A, McMahon, C, Kleinhenz, A.L.W, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR.
Science, 367, 2020
|
|
6OS2
 
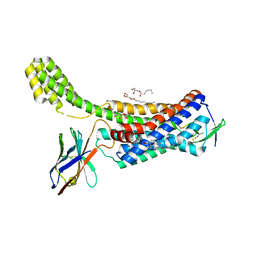 | | Structure of synthetic nanobody-stabilized angiotensin II type 1 receptor bound to TRV026 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Wingler, L.M, Staus, D.P, Skiba, M.A, McMahon, C, Kleinhenz, A.L.W, Lefkowitz, R.J, Kruse, A.C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Angiotensin and biased analogs induce structurally distinct active conformations within a GPCR.
Science, 367, 2020
|
|
6M8R
 
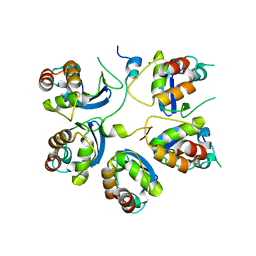 | |
6M8S
 
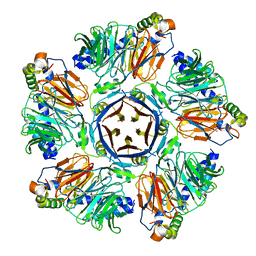 | | Crystal structure of the KCTD12 H1 domain in complex with Gbeta1gamma2 subunits | | Descriptor: | BTB/POZ domain-containing protein KCTD12, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | | Authors: | Zheng, S, Kruse, A.C. | | Deposit date: | 2018-08-22 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.71 Å) | | Cite: | Structural basis for KCTD-mediated rapid desensitization of GABABsignalling.
Nature, 567, 2019
|
|
6MJP
 
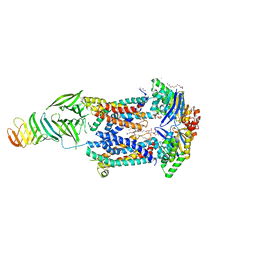 | | LptB(E163Q)FGC from Vibrio cholerae | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, 6-cyclohexylhexyl beta-D-glucopyranoside, ABC transporter ATP-binding protein, ... | | Authors: | Owens, T.W, Kahne, D, Kruse, A.C. | | Deposit date: | 2018-09-21 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of unidirectional export of lipopolysaccharide to the cell surface.
Nature, 567, 2019
|
|
6PL5
 
 | | Structural coordination of polymerization and crosslinking by a peptidoglycan synthase complex | | Descriptor: | Penicillin-binding protein 2/cell division protein FtsI, Peptidoglycan glycosyltransferase RodA, Unknown peptide | | Authors: | Sjodt, M, Rohs, P.D.A, Erlandson, S.C, Zheng, S, Rudner, D.Z, Bernhardt, T.G, Kruse, A.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex.
Nat Microbiol, 5, 2020
|
|
6PL6
 
 | | Structural coordination of polymerization and crosslinking by a peptidoglycan synthase complex | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-amino-2-phenylacetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Penicillin-binding protein 2/cell division protein FtsI, Peptidoglycan glycosyltransferase RodA, ... | | Authors: | Sjodt, M, Rohs, P.D.A, Erlandson, S.C, Zheng, S, Rudner, D.Z, Bernhardt, T.G, Kruse, A.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural coordination of polymerization and crosslinking by a SEDS-bPBP peptidoglycan synthase complex.
Nat Microbiol, 5, 2020
|
|
