2AK5
 
 | | beta PIX-SH3 complexed with a Cbl-b peptide | | Descriptor: | 8-residue peptide from a signal transduction protein CBL-B, Rho guanine nucleotide exchange factor 7 | | Authors: | Jozic, D, Cardenes, N, Deribe, Y.L, Moncalian, G, Hoeller, D, Groemping, Y, Dikic, I, Rittinger, K, Bravo, J. | | Deposit date: | 2005-08-03 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Cbl promotes clustering of endocytic adaptor proteins.
Nat.Struct.Mol.Biol., 12, 2005
|
|
8Q7K
 
 | | IRGQ LIR2 peptide in complex with LC3B | | Descriptor: | 1,2-ETHANEDIOL, Immunity-related GTPase family Q protein, Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Misra, M, Bailey, H.J, Pomirska, J, Dikic, I. | | Deposit date: | 2023-08-16 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Tumor immune evasion through IRGQ-directed autophagy
To Be Published
|
|
4MRT
 
 | | Structure of the Phosphopantetheine Transferase Sfp in Complex with Coenzyme A and a Peptidyl Carrier Protein | | Descriptor: | 4'-phosphopantetheinyl transferase sfp, COENZYME A, GLYCEROL, ... | | Authors: | Tufar, P, Rahighi, S, Kraas, F.I, Kirchner, D.K, Loehr, F, Henrich, E, Koepke, J, Dikic, I, Guentert, P, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2013-09-17 | | Release date: | 2014-04-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of a PCP/Sfp Complex Reveals the Structural Basis for Carrier Protein Posttranslational Modification.
Chem.Biol., 21, 2014
|
|
5M93
 
 | |
5JW7
 
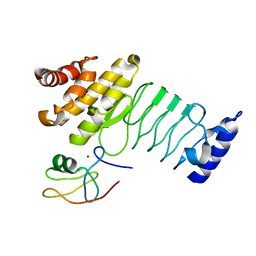 | | Crystal structure of SopA-Trim56 complex | | Descriptor: | E3 ubiquitin-protein ligase SopA, E3 ubiquitin-protein ligase TRIM56, ZINC ION | | Authors: | Bhogaraju, S, Dikic, I. | | Deposit date: | 2016-05-11 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.849 Å) | | Cite: | Structural basis for the recognition and degradation of host TRIM proteins by Salmonella effector SopA.
Nat Commun, 8, 2017
|
|
3F89
 
 | | NEMO CoZi domain | | Descriptor: | NF-kappa-B essential modulator | | Authors: | Rahighi, S, Ikeda, F, Kawasaki, M, Akutsu, M, Suzuki, N, Kato, R, Kensche, T, Uejima, T, Bloor, S, Komander, D, Randow, F, Wakatsuki, S, Dikic, I. | | Deposit date: | 2008-11-11 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation
Cell(Cambridge,Mass.), 136, 2009
|
|
6N6R
 
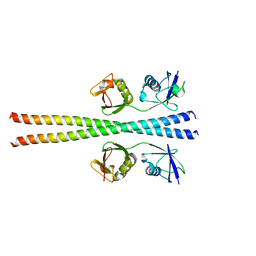 | |
6N6S
 
 | | Crystal structure of ABIN-1 UBAN | | Descriptor: | TNFAIP3-interacting protein 1 | | Authors: | Rahighi, S, Dikic, I, Wakatsuki, S. | | Deposit date: | 2018-11-27 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular Recognition of M1-Linked Ubiquitin Chains by Native and Phosphorylated UBAN Domains.
J.Mol.Biol., 431, 2019
|
|
6N5M
 
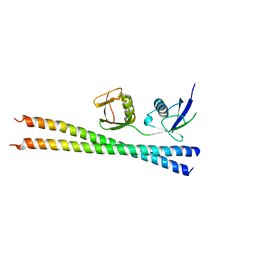 | |
6YVA
 
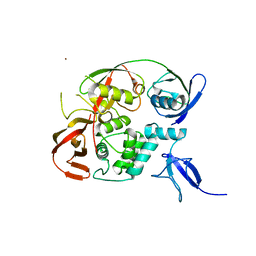 | | PLpro-C111S with mISG15 | | Descriptor: | Replicase polyprotein 1a, Ubiquitin-like protein ISG15, ZINC ION | | Authors: | Shin, D, Dikic, I. | | Deposit date: | 2020-04-28 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity.
Nature, 587, 2020
|
|
2R2Y
 
 | |
4EMO
 
 | | Crystal structure of the PH domain of SHARPIN | | Descriptor: | Sharpin | | Authors: | Stieglitz, B, Haire, L.F, Dikic, I, Rittinger, K. | | Deposit date: | 2012-04-12 | | Release date: | 2012-05-02 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Analysis of SHARPIN, a Subunit of a Large Multi-protein E3 Ubiquitin Ligase, Reveals a Novel Dimerization Function for the Pleckstrin Homology Superfold.
J.Biol.Chem., 287, 2012
|
|
4ZDV
 
 | |
8Q6Q
 
 | |
6G0C
 
 | | Crystal structure of SdeA catalytic core | | Descriptor: | 1,2-ETHANEDIOL, 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, Ubiquitinating/deubiquitinating enzyme SdeA | | Authors: | Kalayil, S, Bhogaraju, S, Basquin, J, Dikic, I. | | Deposit date: | 2018-03-17 | | Release date: | 2018-05-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Insights into catalysis and function of phosphoribosyl-linked serine ubiquitination.
Nature, 557, 2018
|
|
4P09
 
 | | Crystal structure of HOIP PUB domain | | Descriptor: | E3 ubiquitin-protein ligase RNF31 | | Authors: | Akutsu, M, Schaeffer, V, Olma, M.H, Gomes, L.C, Kawasaki, M, Dikic, I. | | Deposit date: | 2014-02-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of OTULIN to the PUB domain of HOIP controls NF-kappa B signaling.
Mol.Cell, 54, 2014
|
|
4P0A
 
 | | Crystal structure of HOIP PUB domain in complex with p97 PIM | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Transitional endoplasmic reticulum ATPase | | Authors: | Akutsu, M, Schaeffer, V, Olma, M.H, Gomes, L.C, Kawasaki, M, Dikic, I. | | Deposit date: | 2014-02-20 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3001 Å) | | Cite: | Binding of OTULIN to the PUB domain of HOIP controls NF-kappa B signaling.
Mol.Cell, 54, 2014
|
|
4P0B
 
 | | Crystal structure of HOIP PUB domain in complex with OTULIN PIM | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Ubiquitin thioesterase otulin | | Authors: | Akutsu, M, Schaeffer, V, Olma, M.H, Gomes, L.C, Kawasaki, M, Dikic, I. | | Deposit date: | 2014-02-20 | | Release date: | 2014-05-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7005 Å) | | Cite: | Binding of OTULIN to the PUB domain of HOIP controls NF-kappa B signaling.
Mol.Cell, 54, 2014
|
|
6YK8
 
 | | OTU-like deubiquitinase from Legionella -Lpg2529 | | Descriptor: | PLATINUM (II) ION, Uncharacterized protein | | Authors: | Shin, D, Dikic, I. | | Deposit date: | 2020-04-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Bacterial OTU deubiquitinases regulate substrate ubiquitination upon Legionella infection.
Elife, 9, 2020
|
|
6YEK
 
 | | Crystal structure of human NEMO apo form | | Descriptor: | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma, isoform CRA_b | | Authors: | Garcia-Pardo, J, Akutsu, M, Busse, P, Skenderovic, A, Maculins, T, Dikic, I. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors by Integrating Protein Engineering and Chemical Screening Platforms.
Cell Chem Biol, 27, 2020
|
|
6XX0
 
 | | Crystal structure of NEMO in complex with Ubv-LIN | | Descriptor: | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma, isoform CRA_b, ... | | Authors: | Akutsu, M, Skenderovic, A, Garcia-Pardo, J, Maculins, T, Dikic, I. | | Deposit date: | 2020-01-26 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors by Integrating Protein Engineering and Chemical Screening Platforms.
Cell Chem Biol, 27, 2020
|
|
2L8J
 
 | | GABARAPL-1 NBR1-LIR complex structure | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, NBR1-LIR peptide | | Authors: | Rogov, V.V, Rozenknop, A, Rogova, N.Y, Loehr, F, Guentert, P, Dikic, I, Doetsch, V. | | Deposit date: | 2011-01-17 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of the Interaction of GABARAPL-1 with the LIR Motif of NBR1.
J.Mol.Biol., 410, 2011
|
|
2MD9
 
 | | Solution Structure of an Active Site Mutant Pepitdyl Carrier Protein | | Descriptor: | Tyrocidine synthase 3 | | Authors: | Tufar, P, Rahighi, S, Kraas, F.I, Kirchner, D.K, Loehr, F, Henrich, E, Koepke, J, Dikic, I, Guentert, P, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2013-09-06 | | Release date: | 2014-04-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal Structure of a PCP/Sfp Complex Reveals the Structural Basis for Carrier Protein Posttranslational Modification.
Chem.Biol., 21, 2014
|
|
2J7I
 
 | | ATYPICAL POLYPROLINE RECOGNITION BY THE CMS N-TERMINAL SH3 DOMAIN. CMS:CD2 HETERODIMER | | Descriptor: | CD2-ASSOCIATED PROTEIN, T-CELL SURFACE ANTIGEN CD2 | | Authors: | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | Deposit date: | 2006-10-09 | | Release date: | 2006-11-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Atypical Polyproline Recognition by the Cms N-Terminal Src Homology 3 Domain.
J.Biol.Chem., 281, 2006
|
|
2J6K
 
 | | N-TERMINAL SH3 DOMAIN OF CMS (CD2AP HUMAN HOMOLOG) | | Descriptor: | CD2-ASSOCIATED PROTEIN, SODIUM ION | | Authors: | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | Deposit date: | 2006-09-29 | | Release date: | 2006-10-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Atypical Polyproline Recognition by the Cms N- Terminal SH3 Domain.
J.Biol.Chem., 281, 2006
|
|
