6PIB
 
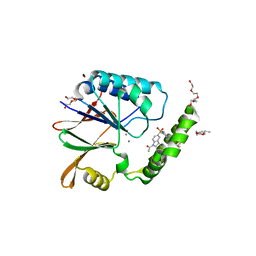 | | Structure of the Klebsiella pneumoniae LpxH-AZ1 complex | | Descriptor: | 1-[5-({4-[3-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)-2,3-dihydro-1H-indol-1-yl]ethan-1-one, MANGANESE (II) ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-26 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5K8K
 
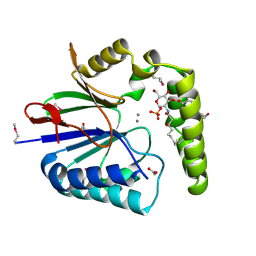 | | Structure of the Haemophilus influenzae LpxH-lipid X complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, ACETATE ION, GLYCEROL, ... | | Authors: | Cho, J, Lee, C.-J, Zhou, P. | | Deposit date: | 2016-05-30 | | Release date: | 2016-08-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the essential Haemophilus influenzae UDP-diacylglucosamine pyrophosphohydrolase LpxH in lipid A biosynthesis.
Nat Microbiol, 1, 2016
|
|
6PH9
 
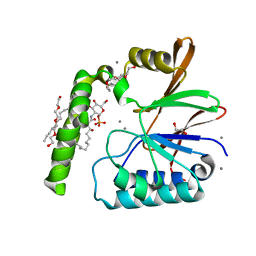 | | Crystal Structure of the Klebsiella pneumoniae LpxH-lipid X complex | | Descriptor: | (R)-((2R,3S,4R,5R,6R)-3-HYDROXY-2-(HYDROXYMETHYL)-5-((R)-3-HYDROXYTETRADECANAMIDO)-6-(PHOSPHONOOXY)TETRAHYDRO-2H-PYRAN-4-YL) 3-HYDROXYTETRADECANOATE, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-25 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PJ3
 
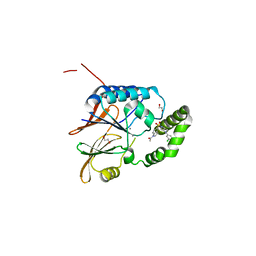 | | Crystal structure of the Klebsiella pneumoniae LpxH/JH-LPH-33 complex | | Descriptor: | 1,2-ETHANEDIOL, 1-[5-({4-[3-chloro-5-(trifluoromethyl)phenyl]piperazin-1-yl}sulfonyl)-2,3-dihydro-1H-indol-1-yl]ethan-1-one, MANGANESE (II) ION, ... | | Authors: | Cho, J, Zhou, P. | | Deposit date: | 2019-06-27 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis of the UDP-diacylglucosamine pyrophosphohydrolase LpxH inhibition by sulfonyl piperazine antibiotics.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2A4V
 
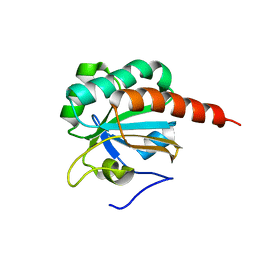 | | Crystal Structure of a truncated mutant of yeast nuclear thiol peroxidase | | Descriptor: | Peroxiredoxin DOT5 | | Authors: | Choi, J, Choi, S, Chon, J.-K, Choi, J, Cha, M.-K, Kim, I.-H, Shin, W. | | Deposit date: | 2005-06-29 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the C107S/C112S mutant of yeast nuclear 2-Cys peroxiredoxin
Proteins, 61, 2005
|
|
218D
 
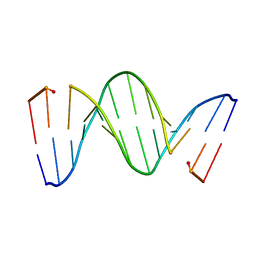 | | THE STRUCTURE OF A NEW CRYSTAL FORM OF A DNA DODECAMER CONTAINING T.(O6ME)G BASE PAIRS | | Descriptor: | DNA (5'-D(*CP*GP*TP*GP*AP*AP*TP*TP*CP*(6OG)P*CP*G)-3') | | Authors: | Vojtechovsky, J, Eaton, M.D, Gaffney, B, Jones, R, Berman, H.M. | | Deposit date: | 1995-07-18 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of a new crystal form of a DNA dodecamer containing T.(O6Me)G base pairs.
Biochemistry, 34, 1995
|
|
1QRI
 
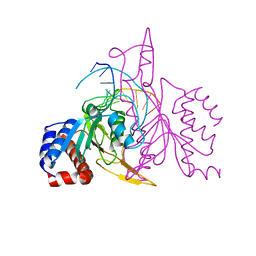 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE COMPLEXES WITH AN E144D MUTATION AT 2.7 A | | Descriptor: | 5'-D(*TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', ECO RI ENDONCULEASE | | Authors: | Choi, J, Kim, Y, Greene, P, Hager, P, Rosenberg, J.M. | | Deposit date: | 1999-06-14 | | Release date: | 1999-06-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structure of the DNA-Eco RI Endonuclease Complexes with the ED144 and RK145 Mutations
To be Published
|
|
1QRH
 
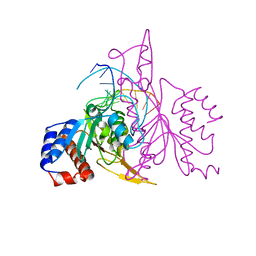 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE COMPLEXES WITH AN R145K MUTATION AT 2.7 A | | Descriptor: | 5'-(TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G*)-3', ECO RI ENDONCULEASE | | Authors: | Choi, J, Kim, Y, Greene, P, Hager, P, Rosenberg, J.M. | | Deposit date: | 1999-06-14 | | Release date: | 1999-06-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Structure of the DNA-Eco RI Endonuclease Complexes with the ED144 and RK145 Mutations
To be Published
|
|
1A6G
 
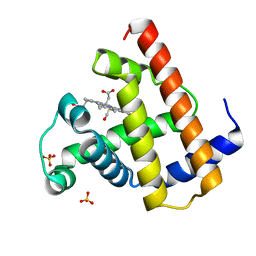 | | CARBONMONOXY-MYOGLOBIN, ATOMIC RESOLUTION | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Vojtechovsky, J, Chu, K, Berendzen, J, Sweet, R.M, Schlichting, I. | | Deposit date: | 1998-02-25 | | Release date: | 1998-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structures of myoglobin-ligand complexes at near-atomic resolution.
Biophys.J., 77, 1999
|
|
1A6M
 
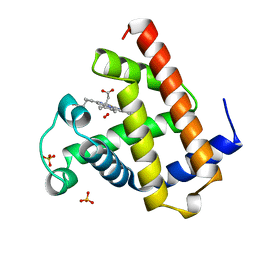 | | OXY-MYOGLOBIN, ATOMIC RESOLUTION | | Descriptor: | MYOGLOBIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Vojtechovsky, J, Chu, K, Berendzen, J, Sweet, R.M, Schlichting, I. | | Deposit date: | 1998-02-26 | | Release date: | 1999-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structures of myoglobin-ligand complexes at near-atomic resolution.
Biophys.J., 77, 1999
|
|
1A6K
 
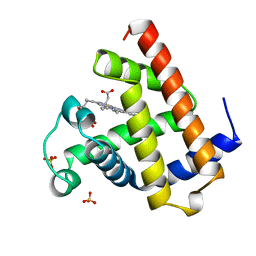 | | AQUOMET-MYOGLOBIN, ATOMIC RESOLUTION | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Vojtechovsky, J, Berendzen, J, Chu, K, Schlichting, I, Sweet, R.M. | | Deposit date: | 1998-02-26 | | Release date: | 1999-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structures of myoglobin-ligand complexes at near-atomic resolution.
Biophys.J., 77, 1999
|
|
1A6N
 
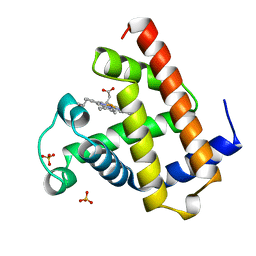 | | DEOXY-MYOGLOBIN, ATOMIC RESOLUTION | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Vojtechovsky, J, Chu, K, Berendzen, J, Sweet, R.M, Schlichting, I. | | Deposit date: | 1998-02-26 | | Release date: | 1999-04-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal structures of myoglobin-ligand complexes at near-atomic resolution.
Biophys.J., 77, 1999
|
|
4TWC
 
 | | 2-Benzamido-N-(1H-benzo[d]imidazol-2-yl)thiazole-4- carboxamide derivatives as potent inhibitors of CK1d/e | | Descriptor: | 2-{[2-(trifluoromethoxy)benzoyl]amino}-N-[6-(trifluoromethyl)-1H-benzimidazol-2-yl]-1,3-thiazole-4-carboxamide, Casein kinase I isoform delta, DIMETHYL SULFOXIDE, ... | | Authors: | Bischof, J, Leban, L, Zaja, M, Grothey, A, Radunsky, B, Othersen, O, Strobl, S, Vitt, D, Knippschild, U. | | Deposit date: | 2014-06-30 | | Release date: | 2014-10-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 2-Benzamido-N-(1H-benzo[d]imidazol-2-yl)thiazole-4-carboxamide derivatives as potent inhibitors of CK1d/e.
Amino Acids, 43, 2012
|
|
4UBR
 
 | |
5OQD
 
 | | PHD2 and winged-helix domain of Polycomblike | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Polycomb protein Pcl, ... | | Authors: | Choi, J, Benda, C, Mueller, J. | | Deposit date: | 2017-08-11 | | Release date: | 2017-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.447 Å) | | Cite: | DNA binding by PHF1 prolongs PRC2 residence time on chromatin and thereby promotes H3K27 methylation.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5Z8T
 
 | | Flavin-containing monooxygenase | | Descriptor: | Acyl-CoA dehydrogenase type 2 domain protein, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Choe, J, Moon, H, Shin, S. | | Deposit date: | 2018-02-01 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structure of putative Flavin-dependent monooxgenase
To Be Published
|
|
6HLL
 
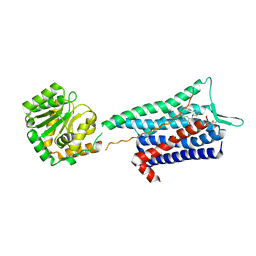 | | Crystal structure of the Neurokinin 1 receptor in complex with the small molecule antagonist CP-99,994 | | Descriptor: | (2~{S},3~{S})-~{N}-[(2-methoxyphenyl)methyl]-2-phenyl-piperidin-3-amine, Substance-P receptor,GlgA glycogen synthase,Substance-P receptor | | Authors: | Schoppe, J, Ehrenmann, J, Klenk, C, Rucktooa, P, Schutz, M, Dore, A.S, Pluckthun, A. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Crystal structures of the human neurokinin 1 receptor in complex with clinically used antagonists.
Nat Commun, 10, 2019
|
|
6HLO
 
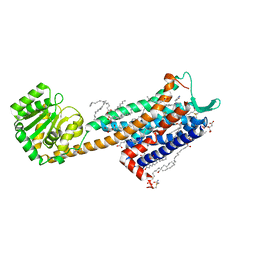 | | Crystal structure of the Neurokinin 1 receptor in complex with the small molecule antagonist Aprepitant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-[[(2~{R},3~{S})-2-[(1~{R})-1-[3,5-bis(trifluoromethyl)phenyl]ethoxy]-3-(4-fluorophenyl)morpholin-4-yl]methyl]-1,2-dihydro-1,2,4-triazol-3-one, CITRIC ACID, ... | | Authors: | Schoppe, J, Ehrenmann, J, Klenk, C, Rucktooa, P, Schutz, M, Dore, A.S, Pluckthun, A. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the human neurokinin 1 receptor in complex with clinically used antagonists.
Nat Commun, 10, 2019
|
|
6HLP
 
 | | Crystal structure of the Neurokinin 1 receptor in complex with the small molecule antagonist Netupitant | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[3,5-bis(trifluoromethyl)phenyl]-~{N},2-dimethyl-~{N}-[4-(2-methylphenyl)-6-(4-methylpiperazin-1-yl)pyridin-3-yl]propanamide, CITRIC ACID, ... | | Authors: | Schoppe, J, Ehrenmann, J, Klenk, C, Rucktooa, P, Schutz, M, Dore, A.S, Pluckthun, A. | | Deposit date: | 2018-09-11 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the human neurokinin 1 receptor in complex with clinically used antagonists.
Nat Commun, 10, 2019
|
|
1QXH
 
 | |
2NLO
 
 | |
2P0M
 
 | | Revised structure of rabbit reticulocyte 15S-lipoxygenase | | Descriptor: | (2E)-3-(2-OCT-1-YN-1-YLPHENYL)ACRYLIC ACID, Arachidonate 15-lipoxygenase, FE (II) ION | | Authors: | Choi, J, Chon, J.K, Kim, S, Shin, W. | | Deposit date: | 2007-02-28 | | Release date: | 2007-10-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational flexibility in mammalian 15S-lipoxygenase: Reinterpretation of the crystallographic data.
Proteins, 70, 2008
|
|
1FAP
 
 | | THE STRUCTURE OF THE IMMUNOPHILIN-IMMUNOSUPPRESSANT FKBP12-RAPAMYCIN COMPLEX INTERACTING WITH HUMAN FRAP | | Descriptor: | FK506-BINDING PROTEIN, FRAP, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Choi, J, Chen, J, Schreiber, S.L, Clardy, J. | | Deposit date: | 1996-03-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP.
Science, 273, 1996
|
|
5IB0
 
 | | PA4534: acetyl CoA complex | | Descriptor: | ACETYL COENZYME *A, GLYCEROL, Uncharacterized protein PA4534 | | Authors: | Choe, J, Shin, S. | | Deposit date: | 2016-02-22 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of PA4534
To Be Published
|
|
3F6F
 
 | |
