1ERI
 
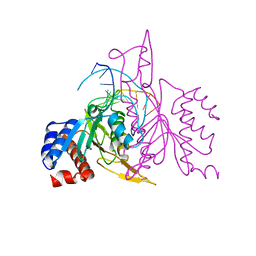 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE-DNA RECOGNITION COMPLEX: THE RECOGNITION NETWORK AND THE INTEGRATION OF RECOGNITION AND CLEAVAGE | | Descriptor: | DNA (5'-D(*TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), PROTEIN (ECO RI ENDONUCLEASE (E.C.3.1.21.4)) | | Authors: | Kim, Y, Grable, J.C, Love, R, Greene, P.J, Rosenberg, J.M. | | Deposit date: | 1994-05-18 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing.
Science, 249, 1990
|
|
1QRI
 
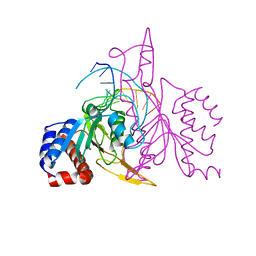 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE COMPLEXES WITH AN E144D MUTATION AT 2.7 A | | Descriptor: | 5'-D(*TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', ECO RI ENDONCULEASE | | Authors: | Choi, J, Kim, Y, Greene, P, Hager, P, Rosenberg, J.M. | | Deposit date: | 1999-06-14 | | Release date: | 1999-06-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | X-Ray Structure of the DNA-Eco RI Endonuclease Complexes with the ED144 and RK145 Mutations
To be Published
|
|
1QRH
 
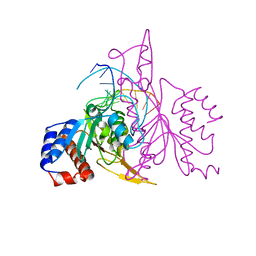 | | X-RAY STRUCTURE OF THE DNA-ECO RI ENDONUCLEASE COMPLEXES WITH AN R145K MUTATION AT 2.7 A | | Descriptor: | 5'-(TP*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G*)-3', ECO RI ENDONCULEASE | | Authors: | Choi, J, Kim, Y, Greene, P, Hager, P, Rosenberg, J.M. | | Deposit date: | 1999-06-14 | | Release date: | 1999-06-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Structure of the DNA-Eco RI Endonuclease Complexes with the ED144 and RK145 Mutations
To be Published
|
|
1SQG
 
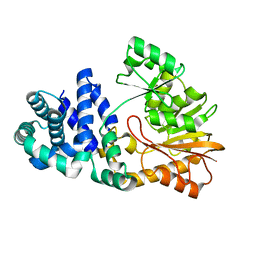 | | The crystal structure of the E. coli Fmu apoenzyme at 1.65 A resolution | | Descriptor: | SUN protein | | Authors: | Foster, P.G, Nunes, C.R, Greene, P, Moustakas, D, Stroud, R.M. | | Deposit date: | 2004-03-18 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The First Structure of an RNA m5C Methyltransferase,
Fmu, Provides Insight into Catalytic Mechanism
and Specific Binding of RNA Substrate
Structure, 11, 2003
|
|
1SQF
 
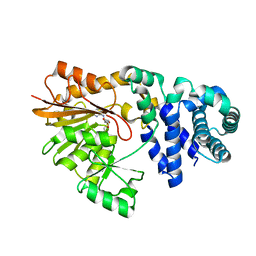 | | The crystal structure of E. coli Fmu binary complex with S-Adenosylmethionine at 2.1 A resolution | | Descriptor: | S-ADENOSYLMETHIONINE, SUN protein | | Authors: | Foster, P.G, Nunes, C.R, Greene, P, Moustakas, D, Stroud, R.M. | | Deposit date: | 2004-03-18 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The First Structure of an RNA m5C Methyltransferase,
Fmu, Provides Insight into Catalytic Mechanism
and Specific Binding of RNA Substrate
Structure, 11, 2003
|
|
4DZT
 
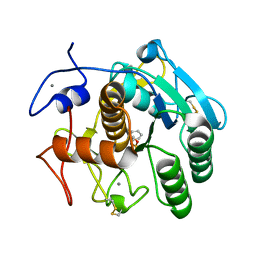 | | Aqualysin I: the crystal structure of a serine protease from an extreme thermophile, Thermus aquaticus YT-1 | | Descriptor: | Aqualysin-1, CALCIUM ION, phenylmethanesulfonic acid | | Authors: | Barnett, B.L, Green, P.R, Strickland, L.C, Oliver, J.D, Rydel, T, Sullivan, J.F. | | Deposit date: | 2012-03-01 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Aqualysin I: the crystal structure of a serine protease from an extreme thermophile, Thermus aquaticus YT-1
To be Published
|
|
