7DNK
 
 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of 5G9 | | Descriptor: | Major capsid protein L1, The heavy chain of 5G9 Fab fragment, The light chain of 5G9 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (6.41 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
7DNL
 
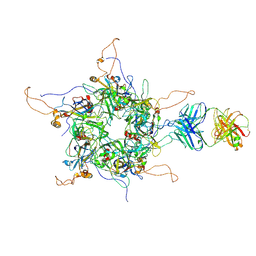 | | 2-fold subparticles refinement of human papillomavirus type 58 pseudovirus in complexed with the Fab fragment of A4B4 | | Descriptor: | Major capsid protein L1, The heavy chain of 2H3 Fab fragment, The light chain of A4B4 Fab fragment | | Authors: | He, M.Z, Chi, X, Zha, Z.H, Zheng, Q.B, Gu, Y, Li, S.W, Xia, N.S. | | Deposit date: | 2020-12-09 | | Release date: | 2020-12-30 | | Last modified: | 2022-12-07 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural basis for the shared neutralization mechanism of three classes of human papillomavirus type 58 antibodies with disparate modes of binding.
J.Virol., 95, 2021
|
|
5I9E
 
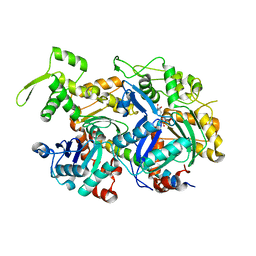 | | Crystal structure of a nuclear actin ternary complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, Actin-related protein 4, ... | | Authors: | Chen, Z, Cao, T. | | Deposit date: | 2016-02-20 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a nuclear actin ternary complex.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3ZMM
 
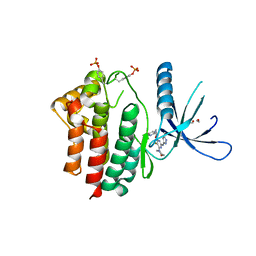 | | Inhibitors of Jak2 Kinase domain | | Descriptor: | 5-FLUORO-4-[(1S)-1-(5-FLUOROPYRIMIDIN-2-YL)ETHOXY]-N-(5-METHYL-1H-PYRAZOL-3-YL)-6-MORPHOLINO-PYRIMIDIN-2-AMINE, ACETYL GROUP, TYROSINE-PROTEIN KINASE JAK2 | | Authors: | Read, J, Green, I, Pollard, H, Howard, T, Mott, R. | | Deposit date: | 2013-02-11 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery of Novel Jak2-Stat Pathway Inhibitors with Extended Residence Time on Target.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4R9V
 
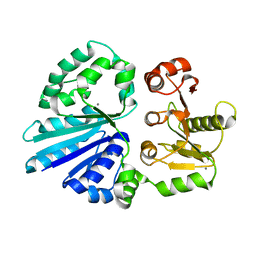 | | Crystal structure of sialyltransferase from photobacterium damselae, residues 113-497 corresponding to the gt-b domain | | Descriptor: | CALCIUM ION, Sialyltransferase 0160 | | Authors: | Li, Y, Huynh, N, Chen, X, Fisher, A.J. | | Deposit date: | 2014-09-08 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
4R83
 
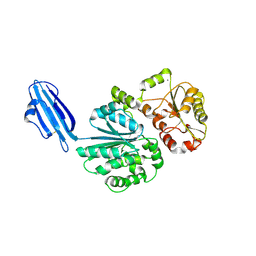 | | Crystal structure of Sialyltransferase from Photobacterium damsela | | Descriptor: | CALCIUM ION, Sialyltransferase 0160 | | Authors: | Fisher, A.J, Chen, X, Li, Y, Huynh, N. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
8UWL
 
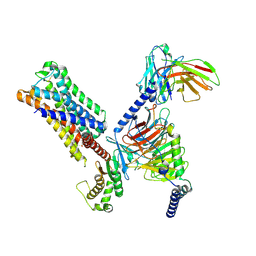 | | 5-HT2AR bound to Lisuride in complex with a mini-Gq protein and an active-state stabilizing single-chain variable fragment (scFv16) obtained by cryo-electron microscopy (cryoEM) | | Descriptor: | 5-hydroxytryptamine receptor 2A, G protein subunit q (Gi2-mini-Gq chimera), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Barros-Alvarez, X, Kim, K, Panova, O, Roth, B.L, Skiniotis, G. | | Deposit date: | 2023-11-06 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | AlphaFold2 structures guide prospective ligand discovery.
Science, 384, 2024
|
|
8V6U
 
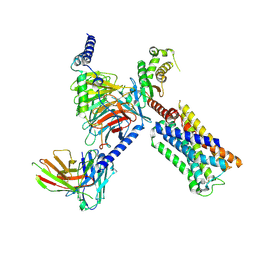 | | 5HT2AR-miniGq heterotrimer in complex with a novel agonist obtained from large scale docking | | Descriptor: | 4-{(3R)-1-[(1R)-1-(pyrimidin-2-yl)ethyl]piperidin-3-yl}phenol, 5-hydroxytryptamine receptor 2A, G protein alpha-subunit q (Gi2-mini-Gq chimera), ... | | Authors: | Gumpper, R.H, Wang, L, Kapolka, N, Skiniotis, G, Roth, B.L. | | Deposit date: | 2023-12-03 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | AlphaFold2 structures guide prospective ligand discovery.
Science, 384, 2024
|
|
4R84
 
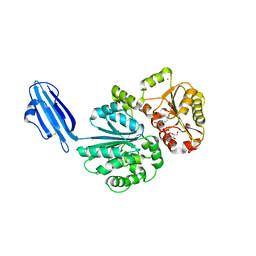 | | Crystal structure of Sialyltransferase from Photobacterium damsela with CMP-3F(a)Neu5Ac bound | | Descriptor: | CALCIUM ION, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, Sialyltransferase 0160 | | Authors: | Fisher, A.J, Chen, X, Li, Y, Huynh, N. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
8ZC3
 
 | | SARS-CoV-2 Omicron BA.4 spike trimer (6P) in complex with 3 D1F6 Fabs (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of D1F6 Fab, ... | | Authors: | Liu, B, Gao, X, Li, Z, Chen, Q, He, J, Xiong, X. | | Deposit date: | 2024-04-28 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | An unconventional VH1-2 antibody tolerates escape mutations and shows an antigenic hotspot on SARS-CoV-2 spike.
Cell Rep, 43, 2024
|
|
8I5I
 
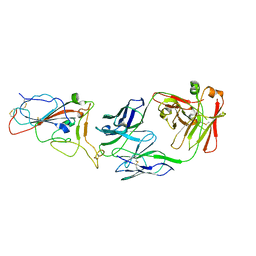 | |
8I5H
 
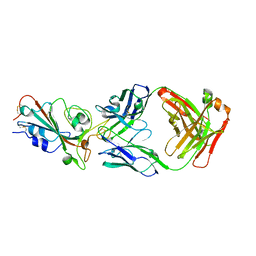 | |
5CRL
 
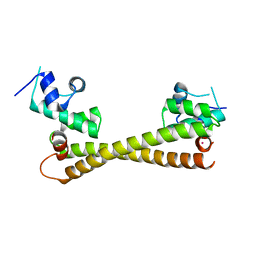 | |
6AZR
 
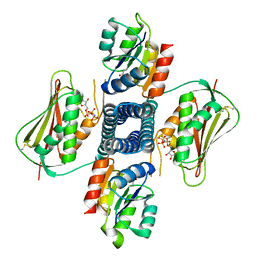 | | Crystal structure of the T264A HK853cp-BeF3-RR468 complex | | Descriptor: | Chemotaxis regulator-transmits chemoreceptor signals to flagelllar motor components CheY, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Rose, J, Zhou, P. | | Deposit date: | 2017-09-11 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.628 Å) | | Cite: | A pH-gated conformational switch regulates the phosphatase activity of bifunctional HisKA-family histidine kinases.
Nat Commun, 8, 2017
|
|
2OGP
 
 | | Solution structure of the second PDZ domain of Par-3 | | Descriptor: | Partitioning-defective 3 homolog | | Authors: | Feng, W, Wu, H, Chen, J, Chan, L.-N, Zhang, M. | | Deposit date: | 2007-01-07 | | Release date: | 2007-12-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | PDZ domains of par-3 as potential phosphoinositide signaling integrators
Mol.Cell, 28, 2007
|
|
7XJP
 
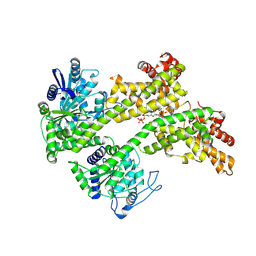 | | Cryo-EM structure of EDS1 and SAG101 with ATP-APDR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5-DIPHOSPHORIBOSE, ISOPROPYL ALCOHOL, ... | | Authors: | Huang, S.J, Jia, A.L, Han, Z.F, Chai, J.J. | | Deposit date: | 2022-04-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity.
Science, 377, 2022
|
|
1AKP
 
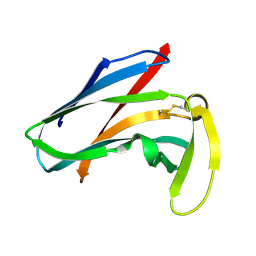 | | SEQUENTIAL 1H,13C AND 15N NMR ASSIGNMENTS AND SOLUTION CONFORMATION OF APOKEDARCIDIN | | Descriptor: | APOKEDARCIDIN | | Authors: | Constantine, K.L, Colson, K.L, Wittekind, M, Friedrichs, M.S, Zein, N, Tuttle, J, Langley, D.R, Leet, J.E, Schroeder, D.R, Lam, K.S, Farmer II, B.T, Metzler, W.J, Bruccoleri, R.E, Mueller, L. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Sequential 1H, 13C, and 15N NMR assignments and solution conformation of apokedarcidin.
Biochemistry, 33, 1994
|
|
1COU
 
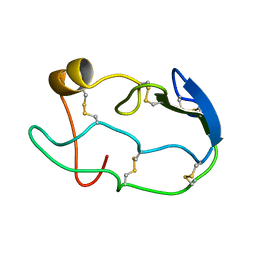 | |
4GRB
 
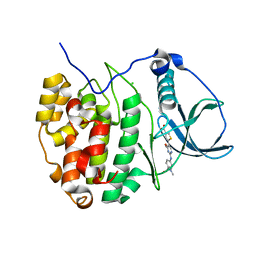 | | Casein kinase 2 (CK2) bound to inhibitor | | Descriptor: | 5-(2-{[4-(dimethylcarbamoyl)phenyl]amino}-4-methoxypyrimidin-5-yl)thiophene-3-carboxylic acid, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Larsen, N.A, Dowling, J.E, Ferguson, A.D. | | Deposit date: | 2012-08-24 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and Property Based Design of Pyrazolo[1,5-a]pyrimidine Inhibitors of CK2 Kinase with Activity in Vivo.
ACS Med Chem Lett, 4, 2013
|
|
6JWE
 
 | | structure of RET G-quadruplex in complex with colchicine | | Descriptor: | DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3'), N-[(7S)-1,2,3,10-tetramethoxy-9-oxo-6,7-dihydro-5H-benzo[d]heptalen-7-yl]ethanamide | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-20 | | Release date: | 2020-02-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
6JWD
 
 | | structure of RET G-quadruplex in complex with berberine | | Descriptor: | BERBERINE, DNA (5'-D(*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*CP*GP*GP*GP*GP*T)-3') | | Authors: | Wang, F, Wang, C, Liu, Y, Lan, W.X, Li, Y.M, Wang, R.X, Cao, C. | | Deposit date: | 2019-04-19 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Colchicine selective interaction with oncogene RET G-quadruplex revealed by NMR.
Chem.Commun.(Camb.), 56, 2020
|
|
7E34
 
 | | Crystal structure of SUN1-Speedy A-CDK2 | | Descriptor: | Cyclin-dependent kinase 2, GLYCEROL, SUN domain-containing protein 1, ... | | Authors: | Chen, Y, Huang, C, Wu, J, Lei, M. | | Deposit date: | 2021-02-08 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The SUN1-SPDYA interaction plays an essential role in meiosis prophase I.
Nat Commun, 12, 2021
|
|
3V5Q
 
 | | Discovery of a selective TRK Inhibitor with efficacy in rodent cancer tumor models | | Descriptor: | 1-(3-{[(3Z)-2-oxo-3-(1H-pyrrol-2-ylmethylidene)-2,3-dihydro-1H-indol-6-yl]amino}phenyl)-3-[3-(trifluoromethyl)phenyl]urea, CHLORIDE ION, NT-3 growth factor receptor | | Authors: | Kreusch, A. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2001 Å) | | Cite: | Discovery of GNF-5837, a Selective TRK Inhibitor with Efficacy in Rodent Cancer Tumor Models.
ACS Med Chem Lett, 3, 2012
|
|
6J1Z
 
 | | WWP2 semi-open conformation | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP2 | | Authors: | Liu, Z.H. | | Deposit date: | 2018-12-30 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A multi-lock inhibitory mechanism for fine-tuning enzyme activities of the HECT family E3 ligases.
Nat Commun, 10, 2019
|
|
6IYO
 
 | |
