3B5Z
 
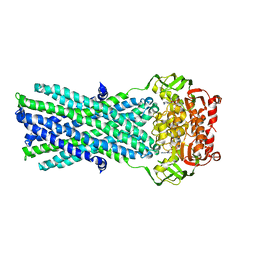 | | Crystal Structure of MsbA from Salmonella typhimurium with ADP Vanadate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lipid A export ATP-binding/permease protein msbA, VANADATE ION | | Authors: | Ward, A, Reyes, C.L, Yu, J, Roth, C.B, Chang, G. | | Deposit date: | 2007-10-26 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Flexibility in the ABC transporter MsbA: Alternating access with a twist.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3FE4
 
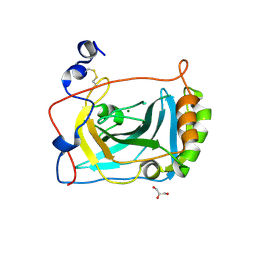 | | Crystal Structure of Human Carbonic Anhydrase vi | | Descriptor: | Carbonic anhydrase 6, GLYCEROL, MAGNESIUM ION | | Authors: | Pilka, E.S, Kochan, G, Krysztofinska, E, Muniz, J, Yue, W.W, Roos, A.K, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-27 | | Release date: | 2008-12-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the secretory isozyme of mammalian carbonic anhydrases CA VI: implications for biological assembly and inhibitor development
Biochem.Biophys.Res.Commun., 419, 2012
|
|
3B5W
 
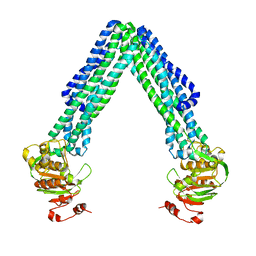 | | Crystal Structure of Eschericia coli MsbA | | Descriptor: | Lipid A export ATP-binding/permease protein msbA | | Authors: | Ward, A, Reyes, C.L, Yu, J, Roth, C.B, Chang, G. | | Deposit date: | 2007-10-26 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.3 Å) | | Cite: | Flexibility in the ABC transporter MsbA: Alternating access with a twist.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7AKZ
 
 | | Deciphering the role of the channel constrictions in the opening mechanism of MexAB-OprM efflux pump from Pseudomonas aeruginosa | | Descriptor: | Outer membrane protein OprM, PALMITIC ACID, octyl beta-D-glucopyranoside | | Authors: | Ntsogo Enguene, V.Y, Monlezun, L, Ma, M, Garnier, C, Lascombe, M.B, Salem, M, Guenard, S, Plesiat, P, Llanes, C, Phan, G, Broutin, I. | | Deposit date: | 2020-10-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Deciphering the role of the channel constrictions in the opening mechanism of MexAB-OprM efflux pump from Pseudomonas aeruginosa
To Be Published
|
|
2XCI
 
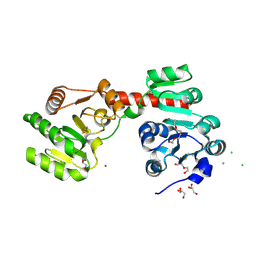 | | Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form | | Descriptor: | 3-DEOXY-D-MANNO-2-OCTULOSONIC ACID TRANSFERASE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Schmidt, H, Hansen, G, Hilgenfeld, R, Mamat, U, Mesters, J.R. | | Deposit date: | 2010-04-26 | | Release date: | 2011-05-11 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Analysis of the Membrane-Embedded Glycosyltransferase Waaa Required for Lipopolysaccharide Synthesis.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3B60
 
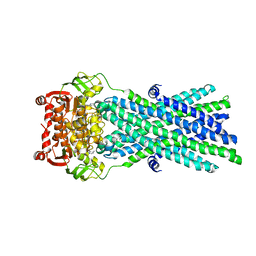 | | Crystal Structure of MsbA from Salmonella typhimurium with AMPPNP, higher resolution form | | Descriptor: | Lipid A export ATP-binding/permease protein msbA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Ward, A, Reyes, C.L, Yu, J, Roth, C.B, Chang, G. | | Deposit date: | 2007-10-26 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Flexibility in the ABC transporter MsbA: Alternating access with a twist.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3B5Y
 
 | | Crystal Structure of MsbA from Salmonella typhimurium with AMPPNP | | Descriptor: | Lipid A export ATP-binding/permease protein msbA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Ward, A, Reyes, C.L, Yu, J, Roth, C.B, Chang, G. | | Deposit date: | 2007-10-26 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Flexibility in the ABC transporter MsbA: Alternating access with a twist.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2WCT
 
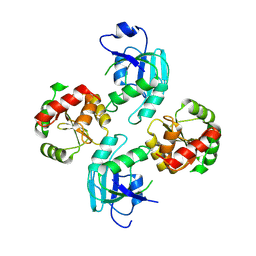 | | human SARS coronavirus unique domain (triclinic form) | | Descriptor: | NON-STRUCTURAL PROTEIN 3 | | Authors: | Tan, J, Vonrhein, C, Smart, O.S, Bricogne, G, Bollati, M, Hansen, G, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2009-03-16 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The Sars-Unique Domain (Sud) of Sars Coronavirus Contains Two Macrodomains that Bind G-Quadruplexes.
Plos Pathog., 5, 2009
|
|
5X8I
 
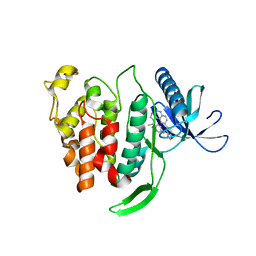 | | Crystal structure of human CLK1 in complex with compound 25 | | Descriptor: | 5-[1-[(1S)-1-(4-fluorophenyl)ethyl]-[1,2,3]triazolo[4,5-c]quinolin-8-yl]-1,3-benzoxazole, Dual specificity protein kinase CLK1 | | Authors: | Sun, Q.Z, Lin, G.F, Li, L.L, Jin, X.T, Huang, L.Y, Zhang, G, Wei, Y.Q, Lu, G.W, Yang, S.Y. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Discovery of Potent and Selective Inhibitors of Cdc2-Like Kinase 1 (CLK1) as a New Class of Autophagy Inducers
J. Med. Chem., 60, 2017
|
|
1JH5
 
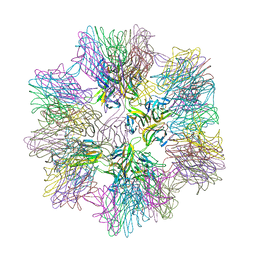 | | Crystal Structure of sTALL-1 of TNF family ligand | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 13B | | Authors: | Liu, Y, Xu, L, Opalka, N, Shu, H.-B, Zhang, G. | | Deposit date: | 2001-06-27 | | Release date: | 2002-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of sTALL-1 reveals a virus-like assembly of TNF family ligands.
Cell(Cambridge,Mass.), 108, 2002
|
|
2LQV
 
 | | YebF | | Descriptor: | Protein yebF | | Authors: | Prehna, G, Zhang, G, Gong, X, Duszyk, M, Okon, M, Mcintosh, L.P, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2012-03-16 | | Release date: | 2012-06-13 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | A Protein Export Pathway Involving Escherichia coli Porins.
Structure, 20, 2012
|
|
5DUJ
 
 | | Crystal structure of ldtMt2 in complex with Faropenem adduct | | Descriptor: | (2R,5R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-5-[(2R)-tetrahydrofuran-2-yl]-2,5-dihydro-1,3-thiazole-4-carboxylic acid, BETA-MERCAPTOETHANOL, L,D-transpeptidase 2, ... | | Authors: | Kumar, P, Lamichhane, G. | | Deposit date: | 2015-09-18 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5DU7
 
 | |
5E1I
 
 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T210 | | Descriptor: | (2S,3R,4R)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-4-(methylsulfanyl)-3,4-dihydro-2H-pyrrole-5-carboxylic acid, GLYCEROL, L,D-transpeptidase 2, ... | | Authors: | Kumar, P, Ginell, S.L, Lamichhane, G. | | Deposit date: | 2015-09-29 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5E51
 
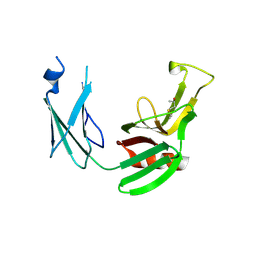 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 1 with Faropenem adduct | | Descriptor: | (3R)-3-hydroxybutanal, L,D-transpeptidase 1 | | Authors: | Kumar, P, Lamichhane, G, Ginell, S.L. | | Deposit date: | 2015-10-07 | | Release date: | 2016-10-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5DVP
 
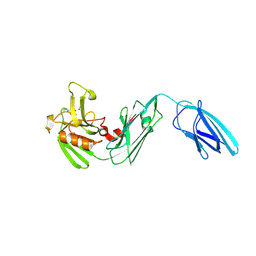 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with Doripenem adduct | | Descriptor: | (2S,3R,4S)-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-4-({(3S,5S)-5-[(sulfamoylamino)methyl]pyrrolidin-3-yl}sulfanyl)-3,4-dihydro-2H-pyrrole-5-carboxylic acid, L,D-transpeptidase 2, PHOSPHONOACETALDEHYDE, ... | | Authors: | Kumar, P, Lamichhane, G. | | Deposit date: | 2015-09-21 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5DZP
 
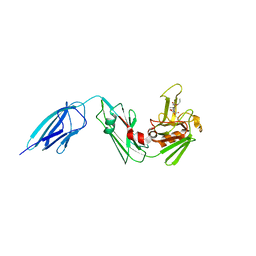 | | Crystal structure of Mycobacterium tuberculosis L,D-transpeptidase 2 with carbapenem drug T206 in conformation B | | Descriptor: | (2~{R},3~{R},4~{R})-4-methyl-3-(2-oxidanylidene-2-propoxy-ethyl)sulfanyl-5-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-2-carboxylic acid, L,D-transpeptidase 2 | | Authors: | Kumar, P, Ginell, S.L, Lamichhane, G. | | Deposit date: | 2015-09-25 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Non-classical transpeptidases yield insight into new antibacterials.
Nat. Chem. Biol., 13, 2017
|
|
5E5L
 
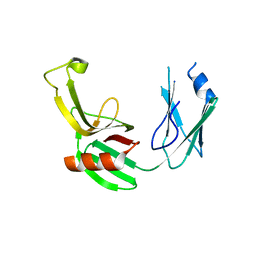 | |
1LVH
 
 | | The Structure of Phosphorylated beta-phosphoglucomutase from Lactoccocus lactis to 2.3 angstrom resolution | | Descriptor: | MAGNESIUM ION, beta-phosphoglucomutase | | Authors: | Lahiri, S.D, Zhang, G, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2002-05-28 | | Release date: | 2002-08-14 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Caught in the act: the structure of phosphorylated beta-phosphoglucomutase from Lactococcus lactis.
Biochemistry, 41, 2002
|
|
2YCQ
 
 | | Crystal structure of checkpoint kinase 2 in complex with inhibitor PV1115 | | Descriptor: | N-{4-[(1E)-N-1H-IMIDAZOL-2-YLETHANEHYDRAZONOYL]PHENYL}-7-NITRO-1H-INDOLE-2-CARBOXAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C.R, Pommier, Y, Shoemaker, R.H, Zhang, G, Waugh, D.S. | | Deposit date: | 2011-03-16 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Characterization of Inhibitor Complexes with Checkpoint Kinase 2 (Chk2), a Drug Target for Cancer Therapy.
J.Struct.Biol., 176, 2011
|
|
8YJG
 
 | |
4ZFQ
 
 | | Structure of M. tuberculosis (3,3) L,D-Transpeptidase, LdtMt5. (Meropenen-adduct form) | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, DI(HYDROXYETHYL)ETHER, L,D-transpeptidase 5 | | Authors: | Basta, L, Ghosh, A, Lamichhane, G, Bianchet, M.A. | | Deposit date: | 2015-04-21 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Loss of a Functionally and Structurally Distinct ld-Transpeptidase, LdtMt5, Compromises Cell Wall Integrity in Mycobacterium tuberculosis.
J.Biol.Chem., 290, 2015
|
|
2YCF
 
 | | Crystal Structure of Checkpoint Kinase 2 in complex with Inhibitor PV1531 | | Descriptor: | (2E)-N-hydroxy-2-[1-(4-{[(4-{(1E)-1-[2-(N'-hydroxycarbamimidoyl)hydrazinylidene]ethyl}phenyl)carbamoyl]amino}phenyl)ethylidene]hydrazinecarboximidamide, MAGNESIUM ION, NITRATE ION, ... | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C.R, Pommier, Y, Shoemaker, R.H, Zhang, G, Waugh, D.S. | | Deposit date: | 2011-03-14 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural Characterization of Inhibitor Complexes with Checkpoint Kinase 2 (Chk2), a Drug Target for Cancer Therapy.
J.Struct.Biol., 176, 2011
|
|
8JLV
 
 | |
2YCS
 
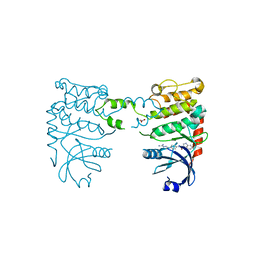 | | Crystal structure of checkpoint kinase 2 in complex with PV788 | | Descriptor: | N-{4-[(1E)-N-CARBAMIMIDOYLETHANEHYDRAZONOYL]PHENYL}-1H-INDOLE-3-CARBOXAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C.R, Pommier, Y, Shoemaker, R.H, Zhang, G, Waugh, D.S. | | Deposit date: | 2011-03-16 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Characterization of Inhibitor Complexes with Checkpoint Kinase 2 (Chk2), a Drug Target for Cancer Therapy.
J.Struct.Biol., 176, 2011
|
|
