4WAU
 
 | | Crystal structure of CENP-M solved by native-SAD phasing | | Descriptor: | Centromere protein M | | Authors: | Weinert, T, Basilico, F, Cecatiello, V, Pasqualato, S, Wang, M. | | Deposit date: | 2014-09-01 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4WAB
 
 | | Crystal structure of mPGES1 solved by native-SAD phasing | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-[(2,2-dimethylpropanoylamino)methyl]phenyl]amino]-1-methyl-6-(2-methyl-2-oxidanyl-propoxy)-N-[2,2,2-tris(fluoranyl)ethyl]benzimidazole-5-carboxamide, GLUTATHIONE, Prostaglandin E synthase,Leukotriene C4 synthase | | Authors: | Weinert, T, Li, D, Howe, N, Caffrey, M, Wang, M. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
5IXY
 
 | | Lactate Dehydrogenase in complex with hydroxylactam inhibitor compound 31: (2~{S})-5-(2-chlorophenyl)sulfanyl-2-(4-morpholin-4-ylphenyl)-4-oxidanyl-2-thiophen-3-yl-1,3-dihydropyridin-6-one | | Descriptor: | (2~{S})-5-(2-chlorophenyl)sulfanyl-2-(4-morpholin-4-ylphenyl)-4-oxidanyl-2-thiophen-3-yl-1,3-dihydropyridin-6-one, L-lactate dehydrogenase A chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Chen, Z, Eigenbrot, C. | | Deposit date: | 2016-03-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cell Active Hydroxylactam Inhibitors of Human Lactate Dehydrogenase with Oral Bioavailability in Mice.
Acs Med.Chem.Lett., 7, 2016
|
|
5JHB
 
 | | Structure of Phosphoinositide 3-kinase gamma (PI3K) bound to the potent inhibitor PIKin3 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, [1-{4-[6-amino-4-(trifluoromethyl)pyridin-3-yl]-6-(morpholin-4-yl)-1,3,5-triazin-2-yl}-3-(chloromethyl)azetidin-3-yl]methanol | | Authors: | Burke, J.E, Inglis, A.J, Williams, R.L. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
5IXS
 
 | | Lactate Dehydrogenase in complex with hydroxylactam inhibitor compound 9: (6R)-3-[(2-chlorophenyl)sulfanyl]-4-hydroxy-6-(3-hydroxyphenyl)-6-(thiophen-3-yl)-5,6-dihydropyridin-2(1H)-one | | Descriptor: | (6R)-3-[(2-chlorophenyl)sulfanyl]-4-hydroxy-6-(3-hydroxyphenyl)-6-(thiophen-3-yl)-5,6-dihydropyridin-2(1H)-one, 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Ultsch, M, Eigenbrot, C. | | Deposit date: | 2016-03-23 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cell Active Hydroxylactam Inhibitors of Human Lactate Dehydrogenase with Oral Bioavailability in Mice.
Acs Med.Chem.Lett., 7, 2016
|
|
5JHA
 
 | | Structure of Phosphoinositide 3-kinase gamma (PI3K) bound to the potent inhibitor PIKin2 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, [1-{4-[6-amino-4-(trifluoromethyl)pyridin-3-yl]-6-(morpholin-4-yl)pyrimidin-2-yl}-3-(chloromethyl)azetidin-3-yl]methanol | | Authors: | Burke, J.E, Inglis, A.J, Williams, R.L. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Deconvolution of Buparlisib's mechanism of action defines specific PI3K and tubulin inhibitors for therapeutic intervention.
Nat Commun, 8, 2017
|
|
2GFX
 
 | |
2GFW
 
 | |
8OUP
 
 | |
2GFY
 
 | |
2GFV
 
 | |
8FLH
 
 | | Bruton's tyrosine kinase in complex with an orthosteric inhibitor | | Descriptor: | 2-(3,5-dichloroanilino)-1-{(3R)-3-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]azepan-1-yl}ethan-1-one, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2022-12-21 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of structural diverse reversible BTK inhibitors utilized to develop a novel in vivo CD69 and CD86 PK/PD mouse model.
Bioorg.Med.Chem.Lett., 80, 2023
|
|
8FLG
 
 | | Bruton's tyrosine kinase in complex with an orthosteric inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, N~2~-(3-chlorophenyl)-N-[(3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]glycinamide, Tyrosine-protein kinase BTK | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2022-12-21 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of structural diverse reversible BTK inhibitors utilized to develop a novel in vivo CD69 and CD86 PK/PD mouse model.
Bioorg.Med.Chem.Lett., 80, 2023
|
|
8FLV
 
 | | Bruton's tyrosine kinase in complex with compound 34 | | Descriptor: | 2-(3,5-dichloroanilino)-1-{(3R)-3-[methyl(7H-pyrrolo[2,3-d]pyrimidin-4-yl)amino]piperidin-1-yl}ethan-1-one, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Metrick, C.M, Marcotte, D.J. | | Deposit date: | 2022-12-22 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of structural diverse reversible BTK inhibitors utilized to develop a novel in vivo CD69 and CD86 PK/PD mouse model.
Bioorg.Med.Chem.Lett., 80, 2023
|
|
7LDJ
 
 | | SARS-CoV-2 receptor binding domain in complex with WNb-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody 2, ... | | Authors: | Pymm, P, Dietrich, M.H, Tan, L.L, Adair, A, Tham, W.H. | | Deposit date: | 2021-01-13 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | SARS-CoV-2 receptor binding domain in complex with WNb-2
Proc.Natl.Acad.Sci.USA, 2021
|
|
7LX5
 
 | |
6UCG
 
 | |
6SSI
 
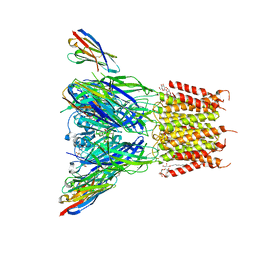 | | Structure of the pentameric ligand-gated ion channel ELIC in complex with a PAM nanobody | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, CALCIUM ION, ... | | Authors: | Ulens, C, Brams, M, Evans, G.L, Spurny, R, Govaerts, C, Pardon, E, Steyaert, J. | | Deposit date: | 2019-09-07 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Modulation of the Erwinia ligand-gated ion channel (ELIC) and the 5-HT 3 receptor via a common vestibule site.
Elife, 9, 2020
|
|
6ZOV
 
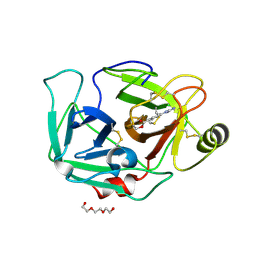 | | ENTEROPEPTIDASE IN COMPLEX WITH COMPOUND 6 | | Descriptor: | 1,2-ETHANEDIOL, 4-carbamimidamidobenzoic acid, Enteropeptidase, ... | | Authors: | Cummings, M.D. | | Deposit date: | 2020-07-07 | | Release date: | 2020-10-21 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Targeting Enteropeptidase with Reversible Covalent Inhibitors To Achieve Metabolic Benefits.
J.Pharmacol.Exp.Ther., 375, 2020
|
|
7MZG
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 42 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, PDI 42 heavy chain, ... | | Authors: | Pymm, P, Chan, L.J, Dietrich, M.H, Tan, L.L, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZK
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 129 and Fab PDI 96 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, ... | | Authors: | Pymm, P, Dietrich, M.H, Tan, L.L, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZH
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 119 | | Descriptor: | Spike protein S1, WCSL 119 heavy chain, WCSL 119 light chain, ... | | Authors: | Pymm, P, Tan, L.L, Dietrich, M.H, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZJ
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 129 and Fab PDI 93 | | Descriptor: | GLYCEROL, PDI 93 heavy chain, PDI 93 light chain, ... | | Authors: | Pymm, P, Dietrich, M.H, Tan, L.L, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZL
 
 | | SARS-CoV-2 receptor binding domain bound to Fab PDI 210 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, PDI 210 heavy chain, PDI 210 light chain, ... | | Authors: | Pymm, P, Chan, L.J, Dietrich, M.H, Tan, L.L, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
7MZI
 
 | | SARS-CoV-2 receptor binding domain bound to Fab WCSL 129 | | Descriptor: | GLYCEROL, Spike protein S1, TETRAETHYLENE GLYCOL, ... | | Authors: | Pymm, P, Tan, L.L, Dietrich, M.H, Chan, L.J, Tham, W.H. | | Deposit date: | 2021-05-24 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Landscape of human antibody recognition of the SARS-CoV-2 receptor binding domain.
Cell Rep, 37, 2021
|
|
