5Y5W
 
 | |
7FIV
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidA and CidBND1-ND2 from wPip(Tunis) | | Descriptor: | CidA_I gamma/2 protein, CidB_I b/2 protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIW
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidAwMel(ST) and CidBND1-ND2 from wPip(Pel) | | Descriptor: | ULP_PROTEASE domain-containing protein, bacteria factor 4,CidA I(Zeta/1) protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIU
 
 | | Crystal structure of the DUB domain of Wolbachia cytoplasmic incompatibility factor CidB from wMel | | Descriptor: | ULP_PROTEASE domain-containing protein | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIT
 
 | | Crystal structure of Wolbachia cytoplasmic incompatibility factor CidA from wMel | | Descriptor: | bacteria factor 1 | | Authors: | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | Deposit date: | 2021-08-01 | | Release date: | 2022-04-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
8KIH
 
 | | PhmA, a type I diterpene synthase without NST/DTE motif | | Descriptor: | (2Z,6E,10E)-2-fluoro-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraen-1-yl trihydrogen diphosphate, MAGNESIUM ION, diterpene synthase, ... | | Authors: | Zhang, B, Ge, H.M, Zhu, A, Zhang, Y. | | Deposit date: | 2023-08-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biosynthesis of Platelet Activating Factor Antagonist Phomactins Revealing a New Class of Type I Diterpene Synthase
To Be Published
|
|
8KI5
 
 | | PhmA, a type I diterpene synthase without NST/DTE motif | | Descriptor: | (2Z,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-ol, PhmA | | Authors: | Zhang, B, Ge, H.M, Zhu, A, Zhang, Y. | | Deposit date: | 2023-08-22 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Biosynthesis of Platelet Activating Factor Antagonist Phomactins Revealing a New Class of Type I Diterpene Synthase
To Be Published
|
|
7F32
 
 | |
8K5N
 
 | | Discovery of Novel PD-L1 Inhibitors That Induce Dimerization and Degradation of PD-L1 Based on Fragment Coupling Strategy | | Descriptor: | 3-[(1~{S})-1-[6-methoxy-3-methyl-5-[[[(2~{S})-5-oxidanylidenepyrrolidin-2-yl]methylamino]methyl]pyridin-2-yl]oxy-2,3-dihydro-1~{H}-inden-4-yl]-2-methyl-~{N}-[5-[[[(2~{S})-5-oxidanylidenepyrrolidin-2-yl]methylamino]methyl]pyridin-2-yl]benzamide, Programmed cell death 1 ligand 1 | | Authors: | Cheng, Y, Xiao, Y.B. | | Deposit date: | 2023-07-22 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Novel PD-L1 Inhibitors That Induce the Dimerization, Internalization, and Degradation of PD-L1 Based on the Fragment Coupling Strategy.
J.Med.Chem., 66, 2023
|
|
7XOE
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Prefusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
7XOG
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide, ... | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
1Q5X
 
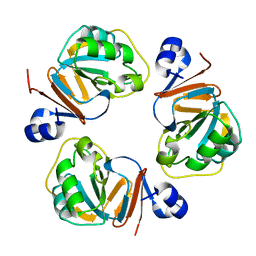 | | Structure of OF RRAA (MENG), a protein inhibitor of RNA processing | | Descriptor: | REGULATOR OF RNASE E ACTIVITY A | | Authors: | Monzingo, A.F, Gao, J, Qiu, J, Georgiou, G, Robertus, J.D. | | Deposit date: | 2003-08-11 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The X-ray Structure of Escherichia coli RraA (MenG), A Protein Inhibitor of RNA Processing.
J.Mol.Biol., 332, 2003
|
|
7CGP
 
 | | Cryo-EM structure of the human mitochondrial translocase TIM22 complex at 3.7 angstrom. | | Descriptor: | 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, Acylglycerol kinase, mitochondrial, ... | | Authors: | Qi, L, Wang, Q, Guan, Z, Yan, C, Yin, P. | | Deposit date: | 2020-07-01 | | Release date: | 2020-10-14 | | Last modified: | 2021-03-17 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the human mitochondrial translocase TIM22 complex.
Cell Res., 31, 2021
|
|
7MOA
 
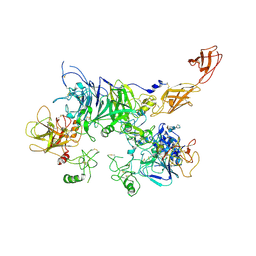 | | Cryo-EM structure of the c-MET II/HGF I complex bound with HGF II in a rigid conformation | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MOB
 
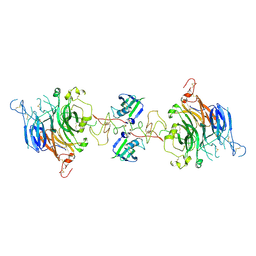 | | Cryo-EM structure of 2:2 c-MET/NK1 complex | | Descriptor: | Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MO8
 
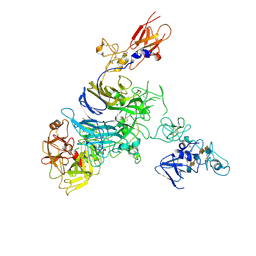 | | Cryo-EM structure of 1:1 c-MET I/HGF I complex after focused 3D refinement of holo-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MO9
 
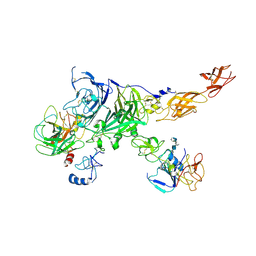 | | Cryo-EM map of the c-MET II/HGF I/HGF II (K4 and SPH) sub-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7MO7
 
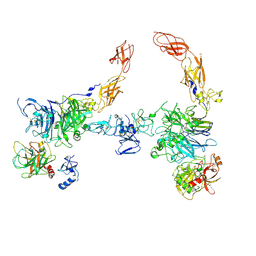 | | Cryo-EM structure of 2:2 c-MET/HGF holo-complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, Hepatocyte growth factor, Hepatocyte growth factor receptor | | Authors: | Uchikawa, E, Chen, Z.M, Xiao, G.Y, Zhang, X.W, Bai, X.C. | | Deposit date: | 2021-05-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of the activation of c-MET receptor.
Nat Commun, 12, 2021
|
|
7CKA
 
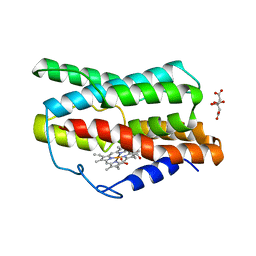 | |
5XGH
 
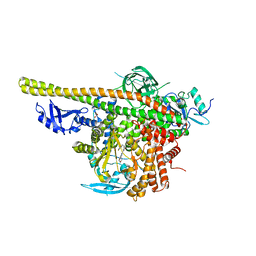 | | Crystal structure of PI3K complex with an inhibitor | | Descriptor: | 3-[(4-fluorophenyl)methylamino]-5-(4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenol, GLYCEROL, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Song, K, Yang, X, Zhao, Y, Jian, Z. | | Deposit date: | 2017-04-13 | | Release date: | 2018-04-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | New Insights into PI3K Inhibitor Design using X-ray Structures of PI3K alpha Complexed with a Potent Lead Compound.
Sci Rep, 7, 2017
|
|
4KDC
 
 | | Crystal Structure of UBIG | | Descriptor: | 3-demethylubiquinone-9 3-methyltransferase | | Authors: | Zhu, Y, Teng, M, Li, X. | | Deposit date: | 2013-04-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and biochemical studies reveal UbiG/Coq3 as a class of novel membrane-binding proteins.
Biochem. J., 470, 2015
|
|
3HIE
 
 | |
2QRV
 
 | | Structure of Dnmt3a-Dnmt3L C-terminal domain complex | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Jia, D, Cheng, X. | | Deposit date: | 2007-07-29 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation.
Nature, 449, 2007
|
|
7K2U
 
 | | DHODH IN COMPLEX WITH LIGAND 13 | | Descriptor: | 6-fluoro-2-(2'-fluoro[1,1'-biphenyl]-4-yl)-N-methoxy-3-methylquinoline-4-carboxamide, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2020-09-09 | | Release date: | 2020-10-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A carboxylic acid isostere screen of the DHODH inhibitor Brequinar.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5XJN
 
 | | cytochrome P450 CREJ in complex with (4-ethylphenyl) dihydrogen phosphate | | Descriptor: | (4-ethylphenyl) dihydrogen phosphate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dong, S, Du, L, Li, S, Feng, Y. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Selective oxidation of aliphatic C-H bonds in alkylphenols by a chemomimetic biocatalytic system
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
