5A2T
 
 | | The Molecular Basis for Flexibility in the Flexible Filamentous Plant Viruses | | Descriptor: | BAMBOO MOSAIC VIRUS, COAT PROTEIN | | Authors: | DiMaio, F, Chen, C.C, Yu, X, Frenz, B, Hsu, Y.H, Lin, N.S, Egelman, E.H. | | Deposit date: | 2015-05-23 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | The Molecular Basis for Flexibility in the Flexible Filamentous Plant Viruses.
Nat.Struct.Mol.Biol., 22, 2015
|
|
3Q98
 
 | | Structure of ygeW encoded protein from E. coli | | Descriptor: | transcarbamylase | | Authors: | Li, Y, Jing, Z, Yu, X, Allewell, N.M, Tuchman, M, Shi, D. | | Deposit date: | 2011-01-07 | | Release date: | 2011-05-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | The ygeW encoded protein from Escherichia coli is a knotted ancestral catabolic transcarbamylase.
Proteins, 79, 2011
|
|
3S6K
 
 | | Crystal structure of xcNAGS | | Descriptor: | Acetylglutamate kinase | | Authors: | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-05-25 | | Release date: | 2012-04-18 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.8018 Å) | | Cite: | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
3S6G
 
 | | Crystal structures of Seleno-substituted mutant mmNAGS in space group P212121 | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, MALONATE ION, ... | | Authors: | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-05-25 | | Release date: | 2012-04-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6681 Å) | | Cite: | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
3S6H
 
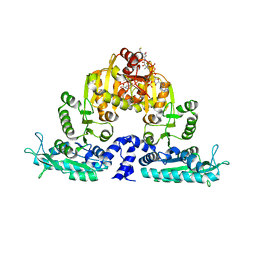 | | Crystal structure of native mmNAGS/k | | Descriptor: | COENZYME A, GLUTAMIC ACID, N-acetylglutamate kinase / N-acetylglutamate synthase | | Authors: | Shi, D, Li, Y, Cabrera-Luque, J, Jin, Z, Yu, X, Allewell, N.M, Tuchman, M. | | Deposit date: | 2011-05-25 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | A Novel N-acetylglutamate synthase architecture revealed by the crystal structure of the bifunctional enzyme from Maricaulis maris.
Plos One, 6, 2011
|
|
8GT6
 
 | | human STING With agonist HB3089 | | Descriptor: | 1-[(2E)-4-{5-carbamoyl-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-[3-(morpholin-4-yl)propoxy]-1H-benzimidazol-1-yl}but-2-en-1-yl]-2-[(1-ethyl-3-methyl-1H-pyrazole-5-carbonyl)amino]-7-methyl-1H-furo[3,2-e]benzimidazole-5-carboxamide, Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
8GSZ
 
 | | Structure of STING SAVI-related mutant V147L | | Descriptor: | Stimulator of interferon genes protein | | Authors: | Wang, Z, Yu, X. | | Deposit date: | 2022-09-07 | | Release date: | 2022-12-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural insights into a shared mechanism of human STING activation by a potent agonist and an autoimmune disease-associated mutation.
Cell Discov, 8, 2022
|
|
6BQ1
 
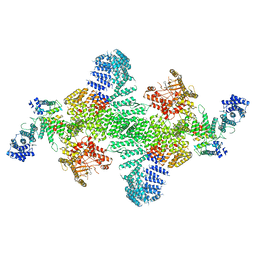 | | Human PI4KIIIa lipid kinase complex | | Descriptor: | 5-{2-amino-1-[4-(morpholin-4-yl)phenyl]-1H-benzimidazol-6-yl}-N-(2-fluorophenyl)-2-methoxypyridine-3-sulfonamide, Phosphatidylinositol 4-kinase III alpha (PI4KA), Protein FAM126A, ... | | Authors: | Lees, J.A, Zhang, Y, Oh, M, Schauder, C.M, Yu, X, Baskin, J, Dobbs, K, Notarangelo, L.D, Camilli, P.D, Walz, T, Reinisch, K.M. | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the human PI4KIII alpha lipid kinase complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6DU3
 
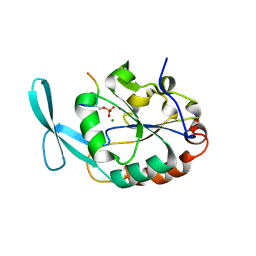 | | Structure of Scp1 D96N bound to REST-pS861/4 peptide | | Descriptor: | Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1, MAGNESIUM ION, REST-pS861 | | Authors: | Burkholder, N.T, Mayfield, J.E, Yu, X, Irani, S, Arce, D.K, Jiang, F, Matthews, W, Xue, Y, Zhang, Y.J. | | Deposit date: | 2018-06-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Phosphatase activity of small C-terminal domain phosphatase 1 (SCP1) controls the stability of the key neuronal regulator RE1-silencing transcription factor (REST).
J. Biol. Chem., 293, 2018
|
|
6DU2
 
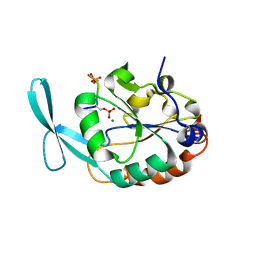 | | Structure of Scp1 D96N bound to REST-pS861/4 peptide | | Descriptor: | MAGNESIUM ION, REST-pS861/4, carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 | | Authors: | Burkholder, N.T, Mayfield, J.E, Yu, X, Irani, S, Arce, D.K, Jiang, F, Matthews, W, Xue, Y, Zhang, Y.J. | | Deposit date: | 2018-06-19 | | Release date: | 2018-09-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Phosphatase activity of small C-terminal domain phosphatase 1 (SCP1) controls the stability of the key neuronal regulator RE1-silencing transcription factor (REST).
J. Biol. Chem., 293, 2018
|
|
7SWD
 
 | | Structure of EBOV GP lacking the mucin-like domain with 1C11 scFv and 1C3 Fab bound | | Descriptor: | 1C11 scFv, 1C3 heavy chain, 1C3 light chain, ... | | Authors: | Milligan, J.C, Yu, X, Saphire, E.O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-04-06 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Asymmetric and non-stoichiometric glycoprotein recognition by two distinct antibodies results in broad protection against ebolaviruses.
Cell, 185, 2022
|
|
4ICZ
 
 | | HER2 1221/1222 phosphorylation regulated by PTPN9 | | Descriptor: | HER2, Tyrosine-protein phosphatase non-receptor type 9 | | Authors: | Xu, Y.F, Wang, H.M, Liu, C.H, Yu, X, Sun, J.P. | | Deposit date: | 2012-12-11 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HER2 1221/1222 phosphorylation regulated by PTPN9
To be Published
|
|
6O3V
 
 | | Crystal structure for RVA-VP3 | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-MONOPHOSPHATE, Protein VP3, ... | | Authors: | Kumar, D, Yu, X, Wang, Z, Hu, L, Prasad, V. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | 2.7 angstrom cryo-EM structure of rotavirus core protein VP3, a unique capping machine with a helicase activity.
Sci Adv, 6, 2020
|
|
4KW8
 
 | |
4KW9
 
 | |
4KW4
 
 | |
4E2I
 
 | | The Complex Structure of the SV40 Helicase Large T Antigen and p68 Subunit of DNA Polymerase Alpha-Primase | | Descriptor: | DNA polymerase alpha subunit B, Large T antigen, ZINC ION | | Authors: | Zhou, B, Arnett, D.R, Yu, X, Brewster, A, Sowd, G.A, Xie, C.L, Vila, S, Gai, D, Fanning, E, Chen, X.S. | | Deposit date: | 2012-03-08 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Structural basis for the interaction of a hexameric replicative helicase with the regulatory subunit of human DNA polymerase alpha-primase.
J.Biol.Chem., 287, 2012
|
|
8EYN
 
 | | Crystal Structure of Human Mitochondrial NADP+ Malic Enzyme 3 in Apo Form | | Descriptor: | CITRIC ACID, NADP-dependent malic enzyme, mitochondrial | | Authors: | Shaffer, P.L, Grell, T.A.J, Mason, M, Thompson, A.A, Riley, D, Wagner, M.V, Steele, R, Ortiz-Meoz, R, Wadia, J, Sharma, S, Yu, X. | | Deposit date: | 2022-10-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
8EYO
 
 | | Crystal Structure of Human Mitochondrial NADP+ Malic Enzyme 3 with NADP bound | | Descriptor: | CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent malic enzyme, ... | | Authors: | Shaffer, P.L, Grell, T.A.J, Mason, M, Thompson, A.A, Riley, D, Wagner, M.V, Steele, R, Ortiz-Meoz, R, Wadia, J, Sharma, S, Yu, X. | | Deposit date: | 2022-10-28 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Integrative structural and functional analysis of human malic enzyme 3: A potential therapeutic target for pancreatic cancer.
Heliyon, 8, 2022
|
|
4GFU
 
 | | PTPN18 in complex with HER2-pY1248 phosphor-peptides | | Descriptor: | HER2-pY1248 phosphor-peptide, Tyrosine-protein phosphatase non-receptor type 18 | | Authors: | Wang, H.M, Yang, F, Du, Y.J, Yang, D.X, Zhang, Y, Yu, X, Sun, J.P. | | Deposit date: | 2012-08-04 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PTPN18-HER2 peptides
To be Published
|
|
7JR7
 
 | | Cryo-EM structure of ABCG5/G8 in complex with Fab 2E10 and 11F4 | | Descriptor: | ATP-binding cassette sub-family G member 5, ATP-binding cassette sub-family G member 8, Fab 11F4 heavy chain, ... | | Authors: | Huang, C.S, Yu, X, Min, X, Wang, Z, Zhang, H. | | Deposit date: | 2020-08-11 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of ABCG5/G8 in complex with modulating antibodies
Commun Biol, 4, 2021
|
|
4GFV
 
 | | PTPN18 in complex with HER2-pY1196 phosphor-peptides | | Descriptor: | HER2-pY1196 phosphor-peptide, Tyrosine-protein phosphatase non-receptor type 18 | | Authors: | Wang, H.M, Yang, F, Du, Y.J, Yang, D.X, Zhang, Y, Yu, X, Sun, J.P. | | Deposit date: | 2012-08-04 | | Release date: | 2013-10-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | PTPN18-HER2 peptides
To be Published
|
|
5ZCJ
 
 | | Crystal structure of complex | | Descriptor: | TP53-binding protein 1, Tudor-interacting repair regulator protein | | Authors: | Wang, J, Yuan, Z, Cui, Y, Xie, R, Wang, M, Ma, Y, Yu, X, Liu, X. | | Deposit date: | 2018-02-17 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Crystal structure of complex
To Be Published
|
|
5U0P
 
 | | Cryo-EM structure of the transcriptional Mediator | | Descriptor: | Mediator complex subunit 10, Mediator complex subunit 11, Mediator complex subunit 14, ... | | Authors: | Tsai, K.-L, Yu, X, Gopalan, S, Chao, T.-C, Zhang, Y, Florens, L, Washburn, M.P, Murakami, K, Conaway, R.C, Conaway, J.W, Asturias, F. | | Deposit date: | 2016-11-26 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mediator structure and rearrangements required for holoenzyme formation.
Nature, 544, 2017
|
|
5VWG
 
 | |
