5KWR
 
 | | Crystal structure of rat Cerebellin-1 | | Descriptor: | Cerebellin-1, alpha-L-fucopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Cheng, S, Ozkan, E. | | Deposit date: | 2016-07-18 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Conformational Plasticity in the Transsynaptic Neurexin-Cerebellin-Glutamate Receptor Adhesion Complex.
Structure, 24, 2016
|
|
5HM7
 
 | | The Intracellular domain of Butyrophilin 3A1 protein | | Descriptor: | BETA-MERCAPTOETHANOL, Butyrophilin subfamily 3 member A1, GLYCEROL, ... | | Authors: | Adams, E.J, Gu, S. | | Deposit date: | 2016-01-15 | | Release date: | 2017-07-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Phosphoantigen-induced conformational change of butyrophilin 3A1 (BTN3A1) and its implication on V gamma 9V delta 2 T cell activation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4O6D
 
 | | West Nile Virus Non-structural protein 1 (NS1) Form 1 crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NS1, ... | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2013-12-20 | | Release date: | 2014-02-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5936 Å) | | Cite: | Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system.
Science, 343, 2014
|
|
4O6B
 
 | |
4O6C
 
 | |
4XHQ
 
 | |
4GNK
 
 | | Crystal structure of Galphaq in complex with full-length human PLCbeta3 | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Lyon, A.M, Tesmer, J.J.G. | | Deposit date: | 2012-08-17 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Full-length G alpha (q)-phospholipase C-beta 3 structure reveals interfaces of the C-terminal coiled-coil domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4H5O
 
 | |
4H5Q
 
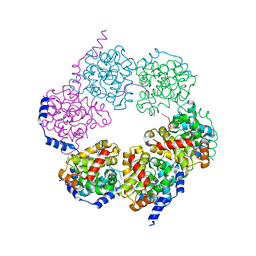 | |
4H5L
 
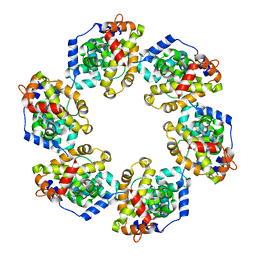 | |
6BDZ
 
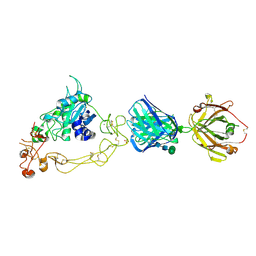 | | ADAM10 Extracellular Domain Bound by the 11G2 Fab | | Descriptor: | 11G2 Fab Heavy Chain, 11G2 Fab Light Chain, CALCIUM ION, ... | | Authors: | Seegar, T.C.M. | | Deposit date: | 2017-10-24 | | Release date: | 2017-12-27 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Regulated Proteolysis by the alpha-Secretase ADAM10.
Cell, 171, 2017
|
|
6BE6
 
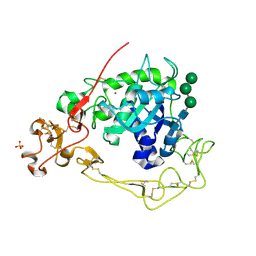 | | ADAM10 Extracellular Domain | | Descriptor: | CALCIUM ION, Disintegrin and metalloproteinase domain-containing protein 10, SULFATE ION, ... | | Authors: | Seegar, T.C.M. | | Deposit date: | 2017-10-24 | | Release date: | 2017-12-27 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Regulated Proteolysis by the alpha-Secretase ADAM10.
Cell, 171, 2017
|
|
6WDP
 
 | | Interleukin 12 receptor subunit beta-1 | | Descriptor: | GLYCEROL, Interleukin-12 receptor subunit beta-1, SULFATE ION | | Authors: | Spangler, J.B, Thomas, C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2020-04-01 | | Release date: | 2021-02-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells.
Cell, 184, 2021
|
|
6WDQ
 
 | | IL23/IL23R/IL12Rb1 signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1, ... | | Authors: | Jude, K.M, Ely, L.K, Glassman, C.R, Thomas, C, Spangler, J.B, Lupardus, P.J, Garcia, K.C. | | Deposit date: | 2020-04-01 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells.
Cell, 184, 2021
|
|
6WGT
 
 | | Crystal structure of HTR2A with hallucinogenic agonist | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562 fusion, CHOLESTEROL, ... | | Authors: | Kim, K.L, Che, T, Krumm, B.E, Roth, B.L. | | Deposit date: | 2020-04-06 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a Hallucinogen-Activated Gq-Coupled 5-HT 2A Serotonin Receptor
Cell(Cambridge,Mass.), 182, 2020
|
|
6WH4
 
 | | Crystal structure of HTR2A with inverse agonist | | Descriptor: | 1-methyl-4-[(5~{S})-3-methylsulfanyl-5,6-dihydrobenzo[b][1]benzothiepin-5-yl]piperazine, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562 fusion, CHOLESTEROL, ... | | Authors: | Kim, K.L, Che, T, Krumm, B.E, Roth, B.L. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of a Hallucinogen-Activated Gq-Coupled 5-HT 2A Serotonin Receptor
Cell(Cambridge,Mass.), 182, 2020
|
|
6E29
 
 | |
6E2H
 
 | |
6CRK
 
 | | Heterotrimeric G-protein in complex with an antibody fragment | | Descriptor: | CITRATE ANION, GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Maeda, S, Dawson, R, Kobilka, B. | | Deposit date: | 2018-03-19 | | Release date: | 2018-10-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of an antibody fragment that stabilizes GPCR/G-protein complexes.
Nat Commun, 9, 2018
|
|
6VHF
 
 | | Crystal structure of RbBP5 interacting domain of Cfp1 | | Descriptor: | PHD-type domain-containing protein, ZINC ION | | Authors: | Joshi, M, Couture, J.F. | | Deposit date: | 2020-01-09 | | Release date: | 2020-01-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | A non-canonical monovalent zinc finger stabilizes the integration of Cfp1 into the H3K4 methyltransferase complex COMPASS.
Nucleic Acids Res., 48, 2020
|
|
6VU8
 
 | | Structure of G-alpha-i bound to its chaperone Ric-8A | | Descriptor: | Guanine nucleotide-binding protein G(i) subunit alpha-1, Resistance to inhibitors of cholinesterase 8 homolog A (C. elegans) | | Authors: | Seven, A.B, Hilger, D. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-18 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.14 Å) | | Cite: | Structures of G alpha Proteins in Complex with Their Chaperone Reveal Quality Control Mechanisms.
Cell Rep, 30, 2020
|
|
6VU5
 
 | | Structure of G-alpha-q bound to its chaperone Ric-8A | | Descriptor: | Guanine nucleotide-binding protein G(q) subunit alpha, Resistance to inhibitors of cholinesterase-8A (Ric-8A) | | Authors: | Seven, A.B, Hilger, D. | | Deposit date: | 2020-02-14 | | Release date: | 2020-03-18 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures of G alpha Proteins in Complex with Their Chaperone Reveal Quality Control Mechanisms.
Cell Rep, 30, 2020
|
|
4TPL
 
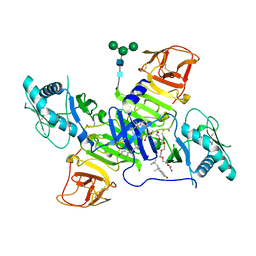 | | West Nile Virus Non-structural protein 1 (NS1) Form 1 crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, OXTOXYNOL-10, ... | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2014-06-08 | | Release date: | 2014-10-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Use of massively multiple merged data for low-resolution S-SAD phasing and refinement of flavivirus NS1.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7T11
 
 | |
7T10
 
 | |
