5VDU
 
 | |
5VDV
 
 | |
5VDT
 
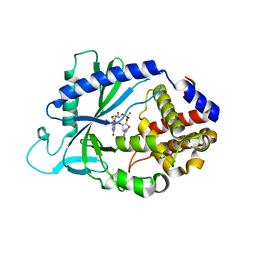 | | Human cyclic GMP-AMP synthase (cGAS) in complex with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Byrnes, L.J, Hall, J.D. | | Deposit date: | 2017-04-03 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.576 Å) | | Cite: | The catalytic mechanism of cyclic GMP-AMP synthase (cGAS) and implications for innate immunity and inhibition.
Protein Sci., 26, 2017
|
|
3MS9
 
 | | ABL kinase in complex with imatinib and a fragment (FRAG1) in the myristate pocket | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, CHLORIDE ION, Tyrosine-protein kinase ABL1, ... | | Authors: | Cowan-Jacob, S.W, Rummel, G, Fendrich, G. | | Deposit date: | 2010-04-29 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding or bending: distinction of allosteric Abl kinase agonists from antagonists by an NMR-based conformational assay.
J.Am.Chem.Soc., 132, 2010
|
|
3MSS
 
 | |
5VDS
 
 | |
5VDW
 
 | | Human cyclic GMP-AMP synthase (cGAS) in complex with Compound F1 | | Descriptor: | Cyclic GMP-AMP synthase, ZINC ION, [2-(1,3-thiazol-4-yl)-1H-benzimidazol-1-yl]acetic acid | | Authors: | Byrnes, L.J, Hall, J.D. | | Deposit date: | 2017-04-03 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | The catalytic mechanism of cyclic GMP-AMP synthase (cGAS) and implications for innate immunity and inhibition.
Protein Sci., 26, 2017
|
|
5VDQ
 
 | |
3QQP
 
 | | Crystal Structure of 11beta-Hydroxysteroid Dehydrogenase 1 (11b-HSD1) in Complex with Urea Inhibitor | | Descriptor: | 3,4-dihydroquinolin-1(2H)-yl[4-(1H-imidazol-5-yl)piperidin-1-yl]methanone, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Loenze, P, Schimanski-Breves, S, Engel, C.K. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Pyrrolidine-pyrazole ureas as potent and selective inhibitors of 11beta-hydroxysteroid-dehydrogenase type 1.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3QM0
 
 | | Crystal structure of RTT109-AC-CoA complex | | Descriptor: | ACETYL COENZYME *A, Histone acetyltransferase RTT109, MERCURY (II) ION | | Authors: | Tang, Y, Marmorstein, R. | | Deposit date: | 2011-02-03 | | Release date: | 2011-02-16 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP.
Nat.Struct.Mol.Biol., 15, 2008
|
|
8RC0
 
 | | Structure of the human 20S U5 snRNP | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schneider, S, Galej, W.P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human 20S U5 snRNP.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1LKT
 
 | | CRYSTAL STRUCTURE OF THE HEAD-BINDING DOMAIN OF PHAGE P22 TAILSPIKE PROTEIN | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Steinbacher, S. | | Deposit date: | 1997-10-17 | | Release date: | 1998-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage.
J.Mol.Biol., 267, 1997
|
|
5OA8
 
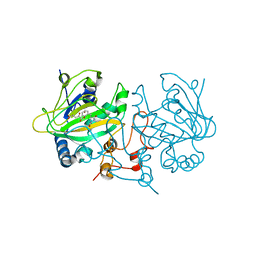 | | Fe(II)/(alpha)ketoglutarate-dependent dioxygenase AsqJ_V72I mutant in complex with demethylated cyclopeptin (1d) | | Descriptor: | 2-OXOGLUTARIC ACID, Iron/alpha-ketoglutarate-dependent dioxygenase asqJ, NICKEL (II) ION, ... | | Authors: | Groll, M, Braeuer, A, Kaila, V.R.I. | | Deposit date: | 2017-06-21 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Catalytic mechanism and molecular engineering of quinolone biosynthesis in dioxygenase AsqJ.
Nat Commun, 9, 2018
|
|
8Q91
 
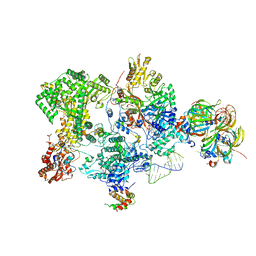 | | Structure of the human 20S U5 snRNP core | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schneider, S, Galej, W.P. | | Deposit date: | 2023-08-19 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the human 20S U5 snRNP.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QAT
 
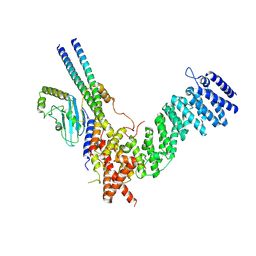 | |
8SHI
 
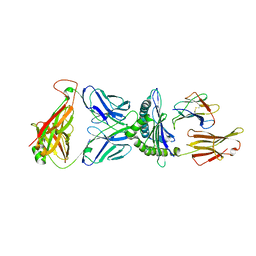 | | Valpha3S1 Vbeta13S1 HLA C 0602 VRSRRCLRL | | Descriptor: | Beta-2-microglobulin, MHC class I antigen (Fragment), T cell receptor alpha, ... | | Authors: | Littler, D.R, Anand, S, Vivian, J.P, Rossjohn, J. | | Deposit date: | 2023-04-14 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (2.90001726 Å) | | Cite: | Complimentary electrostatics dominate T-cell receptor binding to a psoriasis-associated peptide antigen presented by human leukocyte antigen C∗06:02.
J.Biol.Chem., 299, 2023
|
|
8SGD
 
 | | Crystal Structure of CDC3(G) - CDC10(Delta 1-10) heterocomplex from Saccharomyces cerevisiae | | Descriptor: | CDC10 isoform 1, CDC3 isoform 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Saladino, G.C.R, Leonardo, D.A, Pereira, H.M, Garratt, R.C. | | Deposit date: | 2023-04-12 | | Release date: | 2023-06-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | A key piece of the puzzle: The central tetramer of the Saccharomyces cerevisiae septin protofilament and its implications for self-assembly.
J.Struct.Biol., 215, 2023
|
|
5OA7
 
 | | Fe(II)/(alpha)ketoglutarate-dependent dioxygenase AsqJ_V72I mutant in complex with cyclopeptin (1b) | | Descriptor: | 2-OXOGLUTARIC ACID, Iron/alpha-ketoglutarate-dependent dioxygenase asqJ, NICKEL (II) ION, ... | | Authors: | Groll, M, Braeuer, A, Kaila, V.R.I. | | Deposit date: | 2017-06-21 | | Release date: | 2018-04-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Catalytic mechanism and molecular engineering of quinolone biosynthesis in dioxygenase AsqJ.
Nat Commun, 9, 2018
|
|
1JVP
 
 | |
1TZE
 
 | |
2M8O
 
 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in DPC | | Descriptor: | Transmembrane protein gp41 | | Authors: | Serrano, S, Huarte, N, Nieva, J.L, Jimenez, M. | | Deposit date: | 2013-05-23 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Immunogenicity of a Peptide Vaccine, Including the Complete HIV-1 gp41 2F5 Epitope: IMPLICATIONS FOR ANTIBODY RECOGNITION MECHANISM AND IMMUNOGEN DESIGN.
J.Biol.Chem., 289, 2014
|
|
2M8M
 
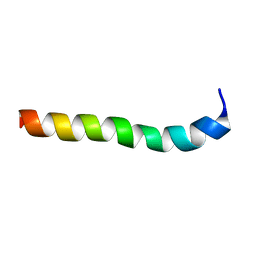 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of hexafluoroisopropanol | | Descriptor: | Transmembrane protein gp41 | | Authors: | Serrano, S, Huarte, N, Nieva, J.L, Jimenez, M. | | Deposit date: | 2013-05-23 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Immunogenicity of a Peptide Vaccine, Including the Complete HIV-1 gp41 2F5 Epitope: IMPLICATIONS FOR ANTIBODY RECOGNITION MECHANISM AND IMMUNOGEN DESIGN.
J.Biol.Chem., 289, 2014
|
|
1YM4
 
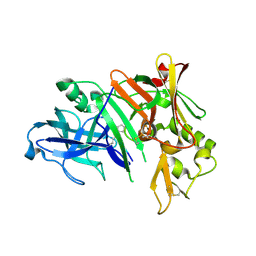 | | Crystal structure of human beta secretase complexed with NVP-AMK640 | | Descriptor: | Beta-secretase 1, NVP-AMK640 INHIBITOR | | Authors: | Hanessian, S, Yun, H, Hou, Y, Yang, G, Bayrakdarian, M, Therrien, E, Moitessier, N, Roggo, S, Veenstra, S. | | Deposit date: | 2005-01-20 | | Release date: | 2006-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based design, synthesis, and memapsin 2 (BACE) inhibitory activity of carbocyclic and heterocyclic peptidomimetics
J.Med.Chem., 48, 2005
|
|
1YM2
 
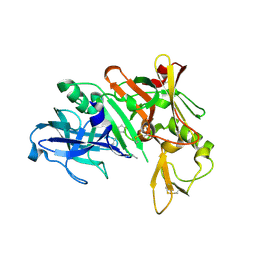 | | Crystal structure of human beta secretase complexed with NVP-AUR200 | | Descriptor: | Beta-secretase 1, NVP-AUR200 INHIBITOR | | Authors: | Hanessian, S, Yun, H, Hou, Y, Yang, G, Bayrakdarian, M, Therrien, E, Moitessier, N, Roggo, S, Veenstra, S. | | Deposit date: | 2005-01-20 | | Release date: | 2006-01-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-based design, synthesis, and memapsin 2 (BACE) inhibitory activity of carbocyclic and heterocyclic peptidomimetics
J.Med.Chem., 48, 2005
|
|
6SZA
 
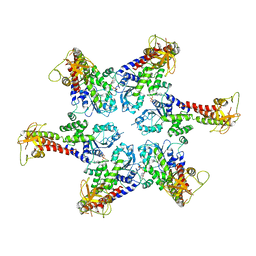 | |
