2C1A
 
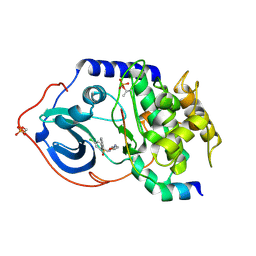 | | Structure of cAMP-dependent protein kinase complexed with Isoquinoline-5-sulfonic acid (2-(2-(4-chlorobenzyloxy)ethylamino) ethyl)amide | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY)ETHYLAMINO)ETHYL)AMIDE | | Authors: | Collins, I, Caldwell, J, Fonseca, T, Donald, A, Bavetsias, V, Hunter, L.J, Garrett, M.D, Rowlands, M.G, Aherne, G.W, Davies, T.G, Berdini, V, Woodhead, S.J, Seavers, L.C.A, Wyatt, P.G, Workman, P, McDonald, E. | | Deposit date: | 2005-09-12 | | Release date: | 2005-11-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-based design of isoquinoline-5-sulfonamide inhibitors of protein kinase B.
Bioorg. Med. Chem., 14, 2006
|
|
4DZT
 
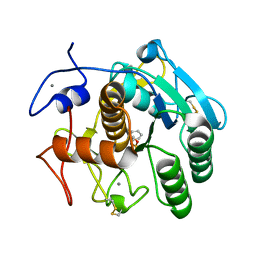 | | Aqualysin I: the crystal structure of a serine protease from an extreme thermophile, Thermus aquaticus YT-1 | | Descriptor: | Aqualysin-1, CALCIUM ION, phenylmethanesulfonic acid | | Authors: | Barnett, B.L, Green, P.R, Strickland, L.C, Oliver, J.D, Rydel, T, Sullivan, J.F. | | Deposit date: | 2012-03-01 | | Release date: | 2012-03-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Aqualysin I: the crystal structure of a serine protease from an extreme thermophile, Thermus aquaticus YT-1
To be Published
|
|
1LVF
 
 | | syntaxin 6 | | Descriptor: | syntaxin 6 | | Authors: | Misura, K.M.S, Bock, J.B, Gonzalez, L.C, Scheller, R.H, Weis, W.I. | | Deposit date: | 2002-05-28 | | Release date: | 2002-07-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structure of the amino-terminal domain of syntaxin 6, a SNAP-25 C homolog.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
8D3B
 
 | |
6A6O
 
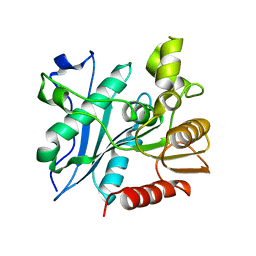 | | Crystal structure of acetyl ester-xyloside bifunctional hydrolase from Caldicellulosiruptor lactoaceticus | | Descriptor: | Esterase/lipase-like protein | | Authors: | Cao, H, Huang, Y, Sun, L.C, Liu, X, Liu, T.F, Wang, F.Z, Xin, F.J. | | Deposit date: | 2018-06-28 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Dual-Substrate Recognition and Catalytic Mechanisms of a Bifunctional Acetyl Ester-Xyloside Hydrolase from Caldicellulosiruptor lactoaceticus.
Acs Catalysis, 9, 2019
|
|
8DB4
 
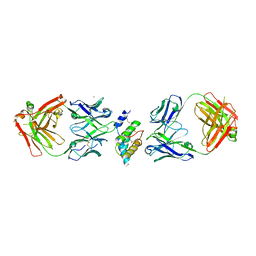 | |
8DFU
 
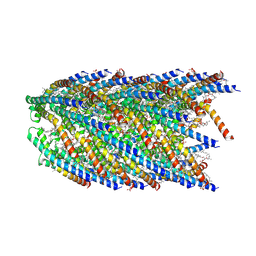 | | Cryo-EM structure of conjugation pili from Aeropyrum pernix | | Descriptor: | (2S)-3-{[(3R,7S,11S,15S)-3,7,11,15,19-pentamethylicosyl]oxy}-2-{[(2R,6S,10S,14R)-2,6,10,14,18-pentamethylnonadecyl]oxy}propyl dihydrogen phosphate, Pilin protein | | Authors: | Beltran, L.C, Egelman, E.H. | | Deposit date: | 2022-06-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery.
Nat Commun, 14, 2023
|
|
8DFT
 
 | | Cryo-EM structure of conjugative pili from Pyrobaculum calidifontis | | Descriptor: | Pilin protein, [(2~{S},7~{S},11~{S},15~{S},19~{R},22~{R},26~{S},30~{R},34~{R},38~{S},43~{S},47~{S},51~{S},55~{R},58~{R},62~{S},66~{R},70~{R})-38-(hydroxymethyl)-7,11,15,19,22,26,30,34,43,47,51,55,58,62,66,70-hexadecamethyl-1,4,37,40-tetraoxacyclodoheptacont-2-yl]methanol | | Authors: | Beltran, L.C, Egelman, E.H. | | Deposit date: | 2022-06-22 | | Release date: | 2023-03-22 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery.
Nat Commun, 14, 2023
|
|
6BBL
 
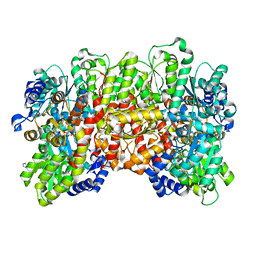 | | Crystal structure of the a-96Gln MoFe protein variant in the presence of the substrate acetylene | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Zadvornyy, O.A, Keable, S.M, Vertemara, J, Eilers, B.J, Karamatullah, D, Rasmussen, A.J, De Gioia, L, Zampella, G, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2017-10-18 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural characterization of the nitrogenase molybdenum-iron protein with the substrate acetylene trapped near the active site.
J. Inorg. Biochem., 180, 2017
|
|
6BKG
 
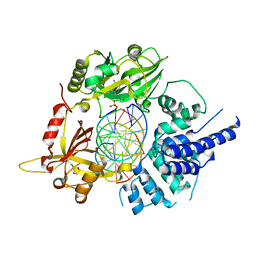 | | Human LigIV catalytic domain with bound DNA-adenylate intermediate in closed conformation | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Moon, A.F, Tumbale, P.P, Schellenberg, M.J, Williams, R.S, Williams, J.G, Kunkel, T.A, Pedersen, L.C, Bebenek, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-07-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structures of DNA-bound human ligase IV catalytic core reveal insights into substrate binding and catalysis.
Nat Commun, 9, 2018
|
|
8E37
 
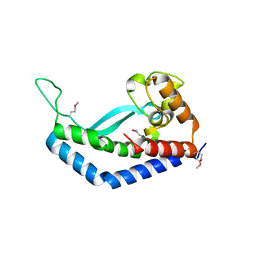 | | Structure of Campylobacter concisus wild-type SeMet PglC | | Descriptor: | N,N'-diacetylbacilliosaminyl-1-phosphate transferase | | Authors: | Vuksanovic, N, Ray, L.C, Imperiali, B, Allen, K.N. | | Deposit date: | 2022-08-16 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Synergistic computational and experimental studies of a phosphoglycosyl transferase membrane/ligand ensemble.
J.Biol.Chem., 299, 2023
|
|
8EXH
 
 | | Agrobacterium tumefaciens Tpilus | | Descriptor: | (14S,17R)-20-amino-17-hydroxy-11,17-dioxo-12,16,18-trioxa-17lambda~5~-phosphaicosan-14-yl tetradecanoate, Protein virB2 | | Authors: | Beltran, L.C, Egelman, E.H. | | Deposit date: | 2022-10-25 | | Release date: | 2023-03-22 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Archaeal DNA-import apparatus is homologous to bacterial conjugation machinery
Nat Commun, 14, 2023
|
|
8EPU
 
 | |
8EPV
 
 | | 2.2 A crystal structure of the lipocalin cat allergen Fel d 7 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Fel d 7 allergen, ... | | Authors: | Min, J, Pedersen, L.C, Geoffrey, M.A. | | Deposit date: | 2022-10-06 | | Release date: | 2023-04-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and ligand binding analysis of the pet allergens Can f 1 and Fel d 7.
Front Allergy, 4, 2023
|
|
6BKF
 
 | | Lysyl-adenylate form of human LigIV catalytic domain with bound DNA substrate in open conformation | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*TP*C)-3'), DNA (5'-D(*GP*TP*CP*CP*GP*AP*CP*GP*AP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Moon, A.F, Tumbale, P.P, Schellenberg, M.J, Williams, R.S, Williams, J.G, Kunkel, T.A, Pedersen, L.C, Bebenek, B. | | Deposit date: | 2017-11-08 | | Release date: | 2018-07-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structures of DNA-bound human ligase IV catalytic core reveal insights into substrate binding and catalysis.
Nat Commun, 9, 2018
|
|
6CDK
 
 | | Characterization of the P1+ intermediate state of nitrogenase P-cluster | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Keable, S.M, Zadvornyy, O.A, Rasmussen, A.J, Danyal, K, Eilers, B.J, Prussia, G.A, LeVan, A.X, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2018-02-08 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the P1+intermediate state of the P-cluster of nitrogenase.
J. Biol. Chem., 293, 2018
|
|
5Z2G
 
 | | Crystal Structure of L-amino acid oxidase from venom of Naja atra | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, L-amino acid oxidase | | Authors: | Kumar, J.V, Chien, K.Y, Wu, W.G, Lin, C.C, Chiang, L.C, Lin, T.H. | | Deposit date: | 2018-01-02 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.676 Å) | | Cite: | Crystal Structure of L-amino acid oxidase from naja atra (Taiwan Cobra)
To Be Published
|
|
6B6I
 
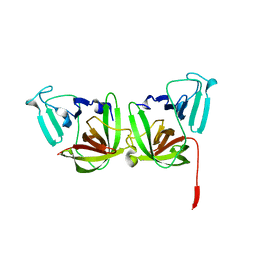 | | 2.4A resolution structure of human Norovirus GII.4 protease | | Descriptor: | 3C-like protease | | Authors: | Muzzarelli, K.M, Kuiper, B.D, Spellmon, N.S, Hackett, J, Brunzelle, J.S, Kovari, I.A, Amblard, F, Yang, Z, Schinazi, R.F, Kovari, L.C. | | Deposit date: | 2017-10-02 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural and Antiviral Studies of the Human Norovirus GII.4 Protease.
Biochemistry, 58, 2019
|
|
1C6V
 
 | | SIV INTEGRASE (CATALYTIC DOMAIN + DNA BIDING DOMAIN COMPRISING RESIDUES 50-293) MUTANT WITH PHE 185 REPLACED BY HIS (F185H) | | Descriptor: | PROTEIN (SIU89134), PROTEIN (SIV INTEGRASE) | | Authors: | Chen, Z, Yan, Y, Munshi, S, Li, Y, Zruygay-Murphy, J, Xu, B, Witmer, M, Felock, P, Wolfe, A, Sardana, V, Emini, E.A, Hazuda, D, Kuo, L.C. | | Deposit date: | 1999-12-21 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50-293)--an initial glance of the viral DNA binding platform.
J.Mol.Biol., 296, 2000
|
|
6WID
 
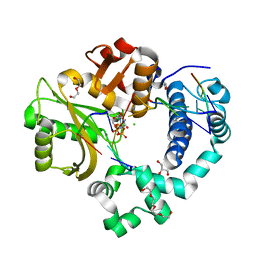 | | Nucleotide incorporation intermediate into quaternary complex of human Polymerase Mu on a complementary DNA double-strand break substrate | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kaminski, A.M, Kunkel, T.A, Pedersen, L.C, Bebenek, K. | | Deposit date: | 2020-04-09 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural snapshots of human DNA polymerase mu engaged on a DNA double-strand break.
Nat Commun, 11, 2020
|
|
6WQ0
 
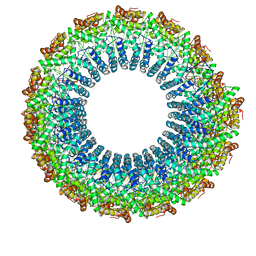 | | Cryo-EM of the S. solfataricus rod-shaped virus, SSRV1 | | Descriptor: | DNA (301-MER), Structural protein | | Authors: | Wang, F, Baquero, D.P, Beltran, L.C, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of filamentous viruses infecting hyperthermophilic archaea explain DNA stabilization in extreme environments.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1BO6
 
 | |
8SLR
 
 | | Crystal Structure of mouse TRAIL | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pedersen, L.C, Xu, D. | | Deposit date: | 2023-04-24 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Heparan sulfate promotes TRAIL-induced tumor cell apoptosis.
Elife, 12, 2024
|
|
1CE1
 
 | | 1.9A STRUCTURE OF THE THERAPEUTIC ANTIBODY CAMPATH-1H FAB IN COMPLEX WITH A SYNTHETIC PEPTIDE ANTIGEN | | Descriptor: | PROTEIN (CAMPATH-1H:HEAVY CHAIN), PROTEIN (CAMPATH-1H:LIGHT CHAIN), PROTEIN (PEPTIDE ANTIGEN) | | Authors: | James, L.C, Hale, G, Waldmann, H, Bloomer, A.C. | | Deposit date: | 1999-03-12 | | Release date: | 1999-06-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 A structure of the therapeutic antibody CAMPATH-1H fab in complex with a synthetic peptide antigen.
J.Mol.Biol., 289, 1999
|
|
1D8M
 
 | | CRYSTAL STRUCTURE OF MMP3 COMPLEXED WITH A HETEROCYCLE-BASED INHIBITOR | | Descriptor: | 1-BENZYL-3-(4-METHOXY-BENZENESULFONYL)-6-OXO-HEXAHYDRO-PYRIMIDINE-4-CARBOXYLIC ACID HYDROXYAMIDE, CALCIUM ION, STROMELYSIN-1 PRECURSOR, ... | | Authors: | Pikul, S, Dunham, K.M, Almstead, N.G, De, B, Natchus, M.G, Taiwo, Y.O, Williams, L.E, Hynd, B.A, Hsieh, L.C, Janusz, M.J, Gu, F, Mieling, G.E. | | Deposit date: | 1999-10-25 | | Release date: | 2000-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Heterocycle-based MMP inhibitors with P2' substituents.
Bioorg.Med.Chem.Lett., 11, 2001
|
|
