6AKK
 
 | | Crystal structure of the second Coiled-coil domain of SIKE1 | | Descriptor: | GLYCEROL, Suppressor of IKBKE 1 | | Authors: | Zhou, L, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
7WHK
 
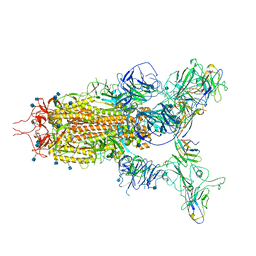 | | The state 3 complex structure of Omicron spike with Bn03 (2-up RBD, 5 nanobodies) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2021-12-30 | | Release date: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7WHI
 
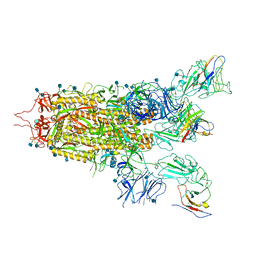 | | The state 2 complex structure of Omicron spike with Bn03 (2-up RBD, 4 nanobodies) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2021-12-30 | | Release date: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
7WHJ
 
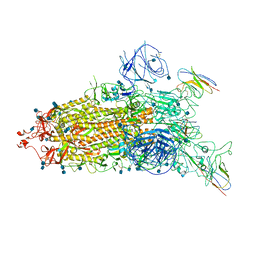 | | The state 1 complex structure of Omicron spike with Bn03 (1-up RBD, 3 nanobodies) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bn03_nano1, Bn03_nano2, ... | | Authors: | Zhan, W.Q, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2021-12-30 | | Release date: | 2022-05-11 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Broad neutralization of SARS-CoV-2 variants by an inhalable bispecific single-domain antibody.
Cell, 185, 2022
|
|
6AKM
 
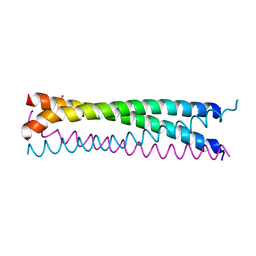 | | Crystal structure of SLMAP-SIKE1 complex | | Descriptor: | GLYCEROL, Sarcolemmal membrane-associated protein, Suppressor of IKBKE 1 | | Authors: | Ma, J, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
5GZB
 
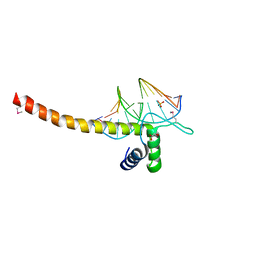 | | Crystal Structure of Transcription Factor TEAD4 in Complex with M-CAT DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*GP*GP*AP*AP*TP*GP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*CP*AP*TP*TP*CP*CP*TP*CP*TP*C)-3'), GLYCEROL, ... | | Authors: | He, F, Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2016-09-28 | | Release date: | 2017-04-19 | | Last modified: | 2017-08-09 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | DNA-binding mechanism of the Hippo pathway transcription factor TEAD4
Oncogene, 36, 2017
|
|
7V2A
 
 | | SARS-CoV-2 Spike trimer in complex with XG014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, The heavy chain of XG014, ... | | Authors: | Wang, K, Wang, X, Pan, L. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7V26
 
 | | XG005-bound SARS-CoV-2 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, XG005 Heavy chain, ... | | Authors: | Zhan, W.Q, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2022-07-06 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
7WKS
 
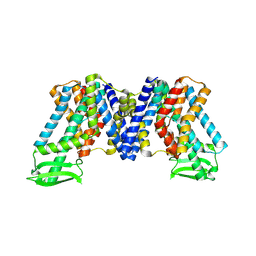 | | Apo state of AtPIN3 | | Descriptor: | Auxin efflux carrier component 3 | | Authors: | Su, N. | | Deposit date: | 2022-01-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
7WKW
 
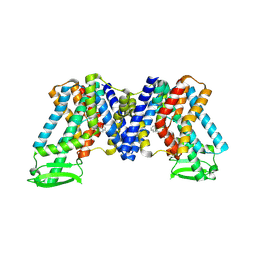 | | NPA bound state of AtPIN3 | | Descriptor: | 2-(naphthalen-1-ylcarbamoyl)benzoic acid, Auxin efflux carrier component 3 | | Authors: | Su, N. | | Deposit date: | 2022-01-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structures and mechanisms of the Arabidopsis auxin transporter PIN3.
Nature, 609, 2022
|
|
7X4E
 
 | | Structure of 10635-DndE | | Descriptor: | DNA sulfur modification protein DndE, GLYCEROL | | Authors: | Haiyan, G, Wei, H, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2022-03-02 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and Functional Analysis of DndE Involved in DNA Phosphorothioation in the Haloalkaliphilic Archaea Natronorubrum bangense JCM10635.
Mbio, 13, 2022
|
|
7EZW
 
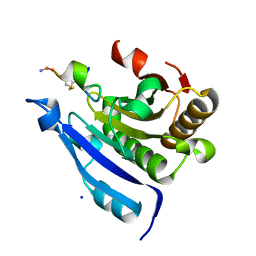 | | Cyclic Peptide that Interacts with the eIF4E Capped-mRNA Binding Site | | Descriptor: | ALA-CYS-GLU-MET-GLY-PHE-PHE-GLN-ASP-CYS-GLY, Eukaryotic translation initiation factor 4E, SODIUM ION | | Authors: | Brown, C.J, Ng, S, Frosi, Y. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Development of a novel peptide aptamer that interacts with the eIF4E capped-mRNA binding site using peptide epitope linker evolution (PELE).
Rsc Chem Biol, 3, 2022
|
|
7F07
 
 | | Autonomous VH domain that interacts with eIF4E at the Capped mRNA Binding site. | | Descriptor: | Eukaryotic translation initiation factor 4E, VH domain (VH-DiFCAP-01) | | Authors: | Brown, C.J, Frosi, Y, Ng, S, Lin, Y.C. | | Deposit date: | 2021-06-03 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Development of a novel peptide aptamer that interacts with the eIF4E capped-mRNA binding site using peptide epitope linker evolution (PELE).
Rsc Chem Biol, 3, 2022
|
|
4Z8M
 
 | | Crystal structure of the MAVS-TRAF6 complex | | Descriptor: | Peptide from Mitochondrial antiviral-signaling protein, TNF receptor-associated factor 6 | | Authors: | Shi, Z.B, Zhou, Z. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Insights into Mitochondrial Antiviral Signaling Protein (MAVS)-Tumor Necrosis Factor Receptor-associated Factor 6 (TRAF6) Signaling
J.Biol.Chem., 290, 2015
|
|
4N6J
 
 | | Crystal structure of human Striatin-3 coiled coil domain | | Descriptor: | Striatin-3 | | Authors: | Chen, C, Shi, Z, Zhou, Z. | | Deposit date: | 2013-10-13 | | Release date: | 2014-02-26 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Striatins contain a noncanonical coiled coil that binds protein phosphatase 2A A subunit to form a 2:2 heterotetrameric core of striatin-interacting phosphatase and kinase (STRIPAK) complex.
J.Biol.Chem., 289, 2014
|
|
4O27
 
 | | Crystal structure of MST3-MO25 complex with WIF motif | | Descriptor: | 5-mer peptide from serine/threonine-protein kinase 24, ADENOSINE-5'-DIPHOSPHATE, Calcium-binding protein 39, ... | | Authors: | Hao, Q, Feng, M, Zhou, Z.C. | | Deposit date: | 2013-12-16 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.185 Å) | | Cite: | Structural insights into regulatory mechanisms of MO25-mediated kinase activation.
J.Struct.Biol., 186, 2014
|
|
4NZW
 
 | | Crystal Structure of STK25-MO25 Complex | | Descriptor: | 5-[(E)-(5-CHLORO-2-OXO-1,2-DIHYDRO-3H-INDOL-3-YLIDENE)METHYL]-N-[2-(DIETHYLAMINO)ETHYL]-2,4-DIMETHYL-1H-PYRROLE-3-CARBOXAMIDE, Calcium-binding protein 39, Serine/threonine-protein kinase 25 | | Authors: | Feng, M, Hao, Q, Zhou, Z.C. | | Deposit date: | 2013-12-13 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.583 Å) | | Cite: | Structural insights into regulatory mechanisms of MO25-mediated kinase activation.
J.Struct.Biol., 186, 2014
|
|
2JYK
 
 | | NMR Structure of a 21 bp DNA duplex preferentially cleaved by Human Topoisomerase II | | Descriptor: | DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DTP*DTP*DAP*DTP*DCP*DAP*DTP*DCP*DGP*DAP*DTP*DCP*DAP*DCP*DGP*DT)-3'), DNA (5'-D(*DAP*DCP*DGP*DTP*DGP*DAP*DTP*DCP*DGP*DAP*DTP*DGP*DAP*DTP*DAP*DAP*DGP*DCP*DTP*DGP*DT)-3') | | Authors: | Masliah, G, Mauffret, O. | | Deposit date: | 2007-12-14 | | Release date: | 2008-07-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Identification of intrinsic dynamics in a DNA sequence preferentially cleaved by topoisomerase II enzyme
J.Mol.Biol., 381, 2008
|
|
4FZF
 
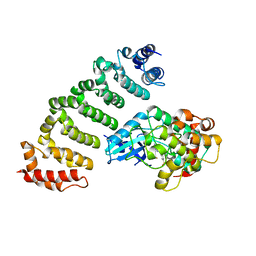 | | Crystal structure of MST4-MO25 complex with DKI | | Descriptor: | 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, Calcium-binding protein 39, Serine/threonine-protein kinase MST4 | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4GEH
 
 | |
4FZD
 
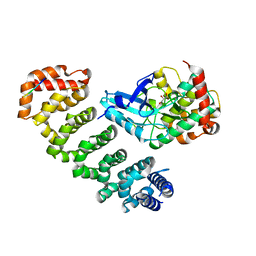 | | Crystal structure of MST4-MO25 complex with WSF motif | | Descriptor: | C-terminal peptide from Serine/threonine-protein kinase MST4, Calcium-binding protein 39, GLYCEROL, ... | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
5YF4
 
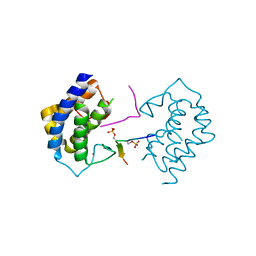 | | A kinase complex MST4-MOB4 | | Descriptor: | MOB-like protein phocein, Peptide from Serine/threonine-protein kinase 26, ZINC ION | | Authors: | Chen, M, Zhou, Z.C. | | Deposit date: | 2017-09-20 | | Release date: | 2018-08-15 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | The MST4-MOB4 complex disrupts the MST1-MOB1 complex in the Hippo-YAP pathway and plays a pro-oncogenic role in pancreatic cancer.
J. Biol. Chem., 293, 2018
|
|
6IUR
 
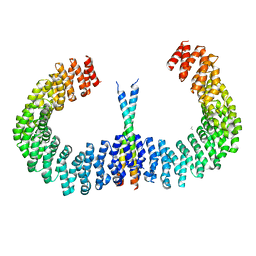 | | A phosphatase complex STRN3-PP2Aa | | Descriptor: | PP2A scaffolding subunit, PROPANE, Striatin-3 | | Authors: | Tang, Y, Zhou, Z.C. | | Deposit date: | 2018-11-30 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Selective Inhibition of STRN3-Containing PP2A Phosphatase Restores Hippo Tumor-Suppressor Activity in Gastric Cancer.
Cancer Cell, 38, 2020
|
|
4FZA
 
 | | Crystal structure of MST4-MO25 complex | | Descriptor: | Calcium-binding protein 39, GLYCEROL, Serine/threonine-protein kinase MST4 | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
7VR6
 
 | | Crystal structure of MlaC from Escherichia coli in quasi-open state | | Descriptor: | 1,2-ETHANEDIOL, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Intermembrane phospholipid transport system binding protein MlaC | | Authors: | Dutta, A, Kanaujia, S.P. | | Deposit date: | 2021-10-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MlaC belongs to a unique class of non-canonical substrate-binding proteins and follows a novel phospholipid-binding mechanism.
J.Struct.Biol., 214, 2022
|
|
