6QB3
 
 | | Apo Mcl1 in a complex with a scFv | | Descriptor: | Induced myeloid leukemia cell differentiation protein Mcl-1, scFv55 | | Authors: | Kazmirski, S, Hargreaves, D. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6QB4
 
 | | Mcl1-scFv complex with an indole acid inhibitor | | Descriptor: | 3-[3-[[(1~{R})-1,2,3,4-tetrahydronaphthalen-1-yl]oxy]propyl]-7-(1,3,5-trimethylpyrazol-4-yl)-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, scFv55 | | Authors: | Hargreaves, D. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6QB9
 
 | | Structure of an anti-Mcl1 scFv | | Descriptor: | L(+)-TARTARIC ACID, scFv55 | | Authors: | Hargreaves, D. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6QBC
 
 | | structure of anti-Mcl1 Fab | | Descriptor: | Anti-Mcl1 Fab Heavy Chain, Anti-Mcl1 Fab Light Chain | | Authors: | Luptak, J. | | Deposit date: | 2018-12-20 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6QF9
 
 | | Structure of an anti-Mcl1 scFv | | Descriptor: | scFv | | Authors: | Luptak, J. | | Deposit date: | 2019-01-09 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6QFC
 
 | | Structure of an anti-Mcl1 scFv | | Descriptor: | DIMETHYL SULFOXIDE, Induced myeloid leukemia cell differentiation protein Mcl-1, scFv55 | | Authors: | Luptak, J. | | Deposit date: | 2019-01-09 | | Release date: | 2019-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Antibody fragments structurally enable a drug-discovery campaign on the cancer target Mcl-1.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
4RAX
 
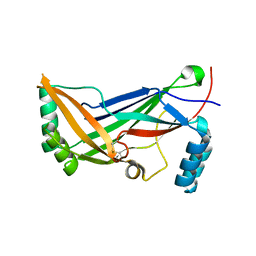 | | A regulatory domain of an ion channel | | Descriptor: | Piezo-type mechanosensitive ion channel component 1 | | Authors: | Ge, J, Yang, M. | | Deposit date: | 2014-09-11 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Architecture of the mammalian mechanosensitive Piezo1 channel.
Nature, 527, 2015
|
|
4U3B
 
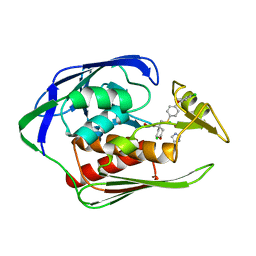 | |
6M67
 
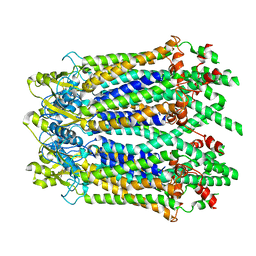 | | The Cryo-EM Structure of Human Pannexin 1 with D376E/D379E Mutation | | Descriptor: | Pannexin-1 | | Authors: | Jin, Q, Bo, Z, Xiang, Z, Xiaokang, Z, Ye, S. | | Deposit date: | 2020-03-13 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of human pannexin 1 channel.
Cell Res., 30, 2020
|
|
4P73
 
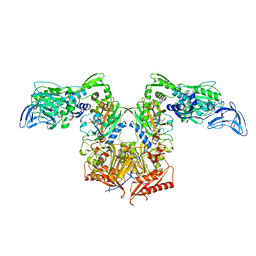 | | PheRS in complex with compound 1a | | Descriptor: | 1-{3-[(4-pyridin-2-ylpiperazin-1-yl)sulfonyl]phenyl}-3-(1,3-thiazol-2-yl)urea, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | The role of a novel auxiliary pocket in bacterial phenylalanyl-tRNA synthetase druggability.
J.Biol.Chem., 289, 2014
|
|
4P75
 
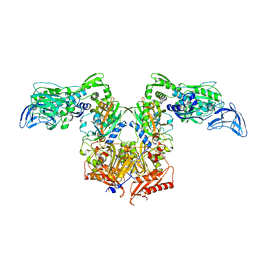 | | PheRS in complex with compound 4a | | Descriptor: | 3-(3-methoxyphenyl)-5-(trifluoromethyl)-1H-pyrazole, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | The role of a novel auxiliary pocket in bacterial phenylalanyl-tRNA synthetase druggability.
J.Biol.Chem., 289, 2014
|
|
4P72
 
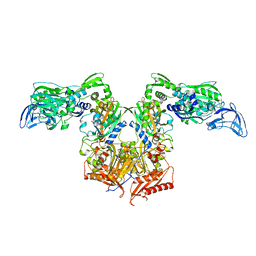 | | PheRS in complex with compound 2a | | Descriptor: | 2-{3-[(4-chloropyridin-2-yl)amino]phenoxy}-N-methylacetamide, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The role of a novel auxiliary pocket in bacterial phenylalanyl-tRNA synthetase druggability.
J.Biol.Chem., 289, 2014
|
|
4P71
 
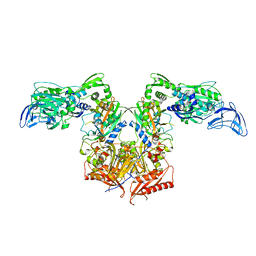 | | Apo PheRS from P. aeuriginosa | | Descriptor: | Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The role of a novel auxiliary pocket in bacterial phenylalanyl-tRNA synthetase druggability.
J.Biol.Chem., 289, 2014
|
|
4P74
 
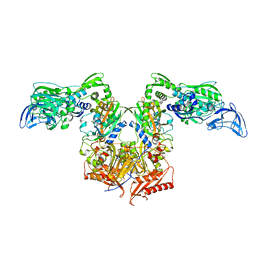 | | PheRS in complex with compound 3a | | Descriptor: | N-[(3S)-1,1-dioxidotetrahydrothiophen-3-yl]-2-[(4-methylphenoxy)methyl]-1,3-thiazole-4-carboxamide, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit | | Authors: | Ferguson, A.D. | | Deposit date: | 2014-03-25 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of a novel auxiliary pocket in bacterial phenylalanyl-tRNA synthetase druggability.
J.Biol.Chem., 289, 2014
|
|
6M68
 
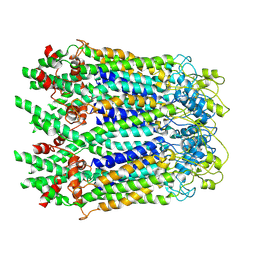 | | The Cryo-EM Structure of Human Pannexin 1 in the Presence of CBX | | Descriptor: | Pannexin-1 | | Authors: | Jin, Q, Bo, Z, Xiang, Z, Xiaokang, Z, Ye, S. | | Deposit date: | 2020-03-13 | | Release date: | 2020-04-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM structures of human pannexin 1 channel.
Cell Res., 30, 2020
|
|
6M66
 
 | | The Cryo-EM Structure of Human Pannexin 1 | | Descriptor: | Pannexin-1 | | Authors: | Jin, Q, Bo, Z, Xiang, Z, Xiaokang, Z, Ye, S. | | Deposit date: | 2020-03-13 | | Release date: | 2020-04-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of human pannexin 1 channel.
Cell Res., 30, 2020
|
|
6LX3
 
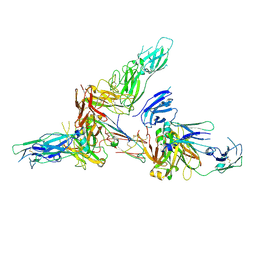 | | Cryo-EM structure of human secretory immunoglobulin A | | Descriptor: | Immunoglobulin J chain, Interleukin-2,Immunoglobulin heavy constant alpha 1, Polymeric immunoglobulin receptor | | Authors: | Wang, Y, Wang, G, Li, Y, Xiao, J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-05-27 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural insights into secretory immunoglobulin A and its interaction with a pneumococcal adhesin.
Cell Res., 30, 2020
|
|
4MCD
 
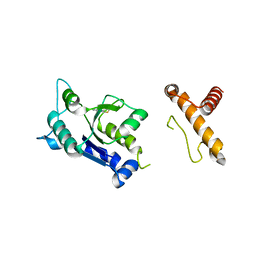 | | hinTrmD in complex with 5-PHENYLTHIENO[2,3-D]PYRIMIDIN-4(3H)-ONE | | Descriptor: | 5-phenylthieno[2,3-d]pyrimidin-4(3H)-one, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Lahiri, S. | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Selective Inhibitors of Bacterial t-RNA-(N(1)G37) Methyltransferase (TrmD) That Demonstrate Novel Ordering of the Lid Domain.
J.Med.Chem., 56, 2013
|
|
4MCC
 
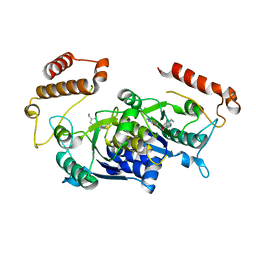 | | HinTrmD in complex with N-[4-(AMINOMETHYL)BENZYL]-4-OXO-3,4-DIHYDROTHIENO[2,3-D]PYRIMIDINE-5-CARBOXAMIDE | | Descriptor: | N-[4-(aminomethyl)benzyl]-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Olivier, N.B, Hill, P. | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Selective Inhibitors of Bacterial t-RNA-(N(1)G37) Methyltransferase (TrmD) That Demonstrate Novel Ordering of the Lid Domain.
J.Med.Chem., 56, 2013
|
|
7S4E
 
 | |
4MCB
 
 | | H.influenzae TrmD in complex with N-(4-{[(1H-IMIDAZOL-2-YLMETHYL)AMINO]METHYL}BENZYL)-4-OXO-3,4-DIHYDROTHIENO[2,3-D]PYRIMIDINE-5-CARBOXAMIDE | | Descriptor: | ACETATE ION, GLYCEROL, N-(4-{[(1H-imidazol-2-ylmethyl)amino]methyl}benzyl)-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidine-5-carboxamide, ... | | Authors: | Olivier, N.B, Hill, P. | | Deposit date: | 2013-08-21 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Selective Inhibitors of Bacterial t-RNA-(N(1)G37) Methyltransferase (TrmD) That Demonstrate Novel Ordering of the Lid Domain.
J.Med.Chem., 56, 2013
|
|
5VFD
 
 | | Diazabicyclooctenone ETX2514 bound to Class D beta lactamase OXA-24 from A. baumannii | | Descriptor: | (2S,5R)-1-formyl-4-methyl-5-[(sulfooxy)amino]-1,2,5,6-tetrahydropyridine-2-carboxamide, (2S,5R)-4-methyl-7-oxo-6-(sulfooxy)-1,6-diazabicyclo[3.2.1]oct-3-ene-2-carboxamide, Beta-lactamase, ... | | Authors: | Olivier, N.B, Lahiri, S. | | Deposit date: | 2017-04-07 | | Release date: | 2017-06-07 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | ETX2514 is a broad-spectrum beta-lactamase inhibitor for the treatment of drug-resistant Gram-negative bacteria including Acinetobacter baumannii.
Nat Microbiol, 2, 2017
|
|
5ID6
 
 | | Structure of Cpf1/RNA Complex | | Descriptor: | Cpf1, MAGNESIUM ION, RNA (5'-R(P*AP*AP*UP*UP*UP*CP*UP*AP*CP*UP*AP*AP*GP*UP*GP*UP*AP*GP*AP*UP*C)-3') | | Authors: | Dong, D, Ren, K, Qiu, X, Wang, J, Huang, Z. | | Deposit date: | 2016-02-24 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | The crystal structure of Cpf1 in complex with CRISPR RNA
Nature, 532, 2016
|
|
3U2K
 
 | | S. aureus GyrB ATPase domain in complex with a small molecule inhibitor | | Descriptor: | 2-chloro-6-(4-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}piperidin-1-yl)pyridine-4-carboxamide, DNA gyrase subunit B, MAGNESIUM ION | | Authors: | Boriack-Sjodin, P.A, Prince, D.B, Eakin, A.E, Sherer, B.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Pyrrolamide DNA gyrase inhibitors: fragment-based nuclear magnetic resonance screening to identify antibacterial agents.
Antimicrob.Agents Chemother., 56, 2012
|
|
3U2D
 
 | | S. aureus GyrB ATPase domain in complex with small molecule inhibitor | | Descriptor: | 4-bromo-5-methyl-N-[1-(3-nitropyridin-2-yl)piperidin-4-yl]-1H-pyrrole-2-carboxamide, DNA gyrase subunit B, MAGNESIUM ION | | Authors: | Boriack-Sjodin, P.A, Prince, D.B, Eakin, A.E, Sherer, B.A. | | Deposit date: | 2011-10-03 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Pyrrolamide DNA gyrase inhibitors: fragment-based nuclear magnetic resonance screening to identify antibacterial agents.
Antimicrob.Agents Chemother., 56, 2012
|
|
