6M66
 
 | | The Cryo-EM Structure of Human Pannexin 1 | | Descriptor: | Pannexin-1 | | Authors: | Jin, Q, Bo, Z, Xiang, Z, Xiaokang, Z, Ye, S. | | Deposit date: | 2020-03-13 | | Release date: | 2020-04-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures of human pannexin 1 channel.
Cell Res., 30, 2020
|
|
7V2A
 
 | | SARS-CoV-2 Spike trimer in complex with XG014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, The heavy chain of XG014, ... | | Authors: | Wang, K, Wang, X, Pan, L. | | Deposit date: | 2021-08-07 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | An ultrapotent pan-beta-coronavirus lineage B ( beta-CoV-B) neutralizing antibody locks the receptor-binding domain in closed conformation by targeting its conserved epitope.
Protein Cell, 13, 2022
|
|
6PAV
 
 | |
8FJB
 
 | |
8FJA
 
 | |
5DZO
 
 | |
6PAU
 
 | |
4Y07
 
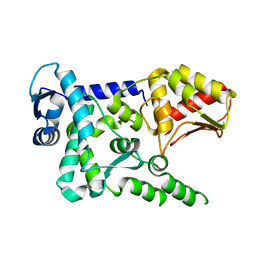 | | Crystal structure of the HECT domain of human WWP2 | | Descriptor: | NEDD4-like E3 ubiquitin-protein ligase WWP2 | | Authors: | Gong, W, Li, J, Li, Z, Xu, Y. | | Deposit date: | 2015-02-05 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.507 Å) | | Cite: | Structure of the HECT domain of human WWP2
Acta Crystallogr.,Sect.F, 71, 2015
|
|
8E2B
 
 | | N-terminal domain of S. aureus GpsB | | Descriptor: | Cell cycle protein GpsB, GLYCEROL | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2022-08-14 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Staphylococcus aureus FtsZ and PBP4 bind to the conformationally dynamic N-terminal domain of GpsB.
Elife, 13, 2024
|
|
7A3O
 
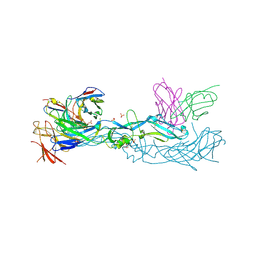 | | Crystal structure of dengue 1 virus envelope glycoprotein in complex with the scFv fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | Core protein, GLYCEROL, SULFATE ION, ... | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3R
 
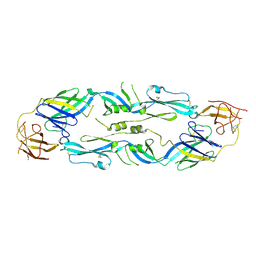 | | Crystal structure of dengue 1 virus envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Core protein, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
8E2C
 
 | |
7A3N
 
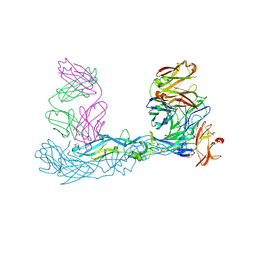 | | Crystal structure of Zika virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | CALCIUM ION, Core protein, EDE1 C10 Fab | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3T
 
 | | Crystal structure of dengue 3 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE1 C8 | | Descriptor: | Core protein, EDE1 C8 antibody Fab fragment, GLYCEROL, ... | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3U
 
 | | Crystal structure of Zika virus envelope glycoprotein in complex with the divalent F(ab')2 fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | EDE1 C10 antibody divalent F(ab')2 fragment, EDE1 C10 divalent F(ab')2 fragment, Envelope protein | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3P
 
 | | Crystal structure of dengue 3 virus envelope glycoprotein in complex with the scFv fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Single Chain Variable Fragment | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3S
 
 | | Crystal structure of dengue 3 virus envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, SULFATE ION | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Navarro-Sanchez, E, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7A3Q
 
 | | Crystal structure of dengue 4 virus envelope glycoprotein in complex with the scFv fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | (2S)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, ... | | Authors: | Sharma, A, Vaney, M.C, Guardado-Calvo, P, Duquerroy, S, Rouvinski, A, Rey, F.A. | | Deposit date: | 2020-08-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The epitope arrangement on flavivirus particles contributes to Mab C10's extraordinary neutralization breadth across Zika and dengue viruses.
Cell, 184, 2021
|
|
7XOE
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Prefusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
7XOG
 
 | | Cryo-EM structure of S glycoprotein encoded by the Covid-19 mRNA vaccine candidate RQ3013 (Postfusion state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,peptide, ... | | Authors: | Wu, Z, Yu, Z, Tan, S, Lu, J, Lu, G, Lin, J. | | Deposit date: | 2022-05-01 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Preclinical evaluation of RQ3013, a broad-spectrum mRNA vaccine against SARS-CoV-2 variants.
Sci Bull (Beijing), 68, 2023
|
|
1AKN
 
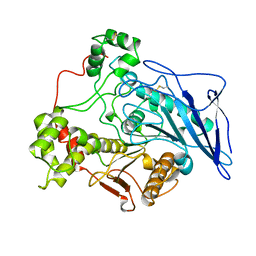 | | STRUCTURE OF BILE-SALT ACTIVATED LIPASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILE-SALT ACTIVATED LIPASE | | Authors: | Wang, X, Zhang, X. | | Deposit date: | 1997-05-23 | | Release date: | 1998-05-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of bovine bile salt activated lipase: insights into the bile salt activation mechanism.
Structure, 5, 1997
|
|
7XLB
 
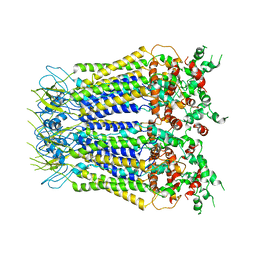 | |
1AQL
 
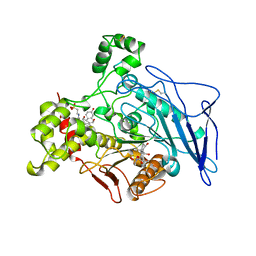 | |
5DZN
 
 | | human T-cell immunoglobulin and mucin domain protein 4 | | Descriptor: | T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Yuan, S, Rao, Z, Wang, X. | | Deposit date: | 2015-09-25 | | Release date: | 2015-11-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | TIM-1 acts a dual-attachment receptor for Ebolavirus by interacting directly with viral GP and the PS on the viral envelope.
Protein Cell, 6, 2015
|
|
4PAR
 
 | | The 5-Hydroxymethylcytosine-Specific Restriction Enzyme AbaSI in a Complex with Product-like DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA 14-MER, DNA 18-MER, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2014-04-09 | | Release date: | 2014-06-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of 5-hydroxymethylcytosine-specific restriction enzyme, AbaSI, in complex with DNA.
Nucleic Acids Res., 42, 2014
|
|
