4KWM
 
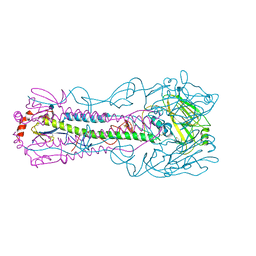 | | Structure of a/anhui/5/2005 h5 ha | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yang, H, Shore, D.A, Carney, P.J, Chang, J.C, Stevens, J. | | Deposit date: | 2013-05-24 | | Release date: | 2014-06-25 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and antigenic variation among diverse clade 2 H5N1 viruses.
Plos One, 8, 2013
|
|
6BCX
 
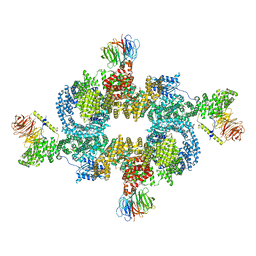 | | mTORC1 structure refined to 3.0 angstroms | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E-binding protein 1, MAGNESIUM ION, ... | | Authors: | Pavletich, N.P, Yang, H. | | Deposit date: | 2017-10-20 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40.
Nature, 552, 2017
|
|
6BCU
 
 | |
8JJ5
 
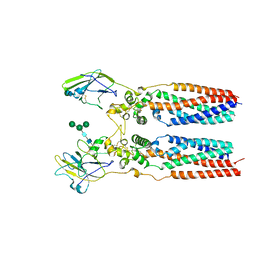 | | Porcine uroplakin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Tetraspanin, ... | | Authors: | Oda, T, Yanagisawa, H, Kikkawa, M. | | Deposit date: | 2023-05-29 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM elucidates the uroplakin complex structure within liquid-crystalline lipids in the porcine urothelial membrane.
Commun Biol, 6, 2023
|
|
5Y8E
 
 | |
5Y8F
 
 | |
6ANV
 
 | | Crystal structure of anti-CRISPR protein AcrF1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, H, Patel, D.J. | | Deposit date: | 2017-08-14 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.265 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
6ANW
 
 | | Crystal structure of anti-CRISPR protein AcrF10 | | Descriptor: | anti-CRISPR protein AcrF10 | | Authors: | Yang, H, Patel, D.J. | | Deposit date: | 2017-08-14 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
8HWN
 
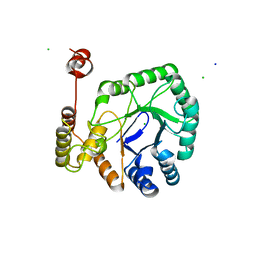 | | aldo-keto reductase DepB | | Descriptor: | CHLORIDE ION, DepB, SODIUM ION | | Authors: | Chen, M, Yang, H, Lu, F. | | Deposit date: | 2022-12-31 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of DepB capable of DON detoxification
To Be Published
|
|
1EQ0
 
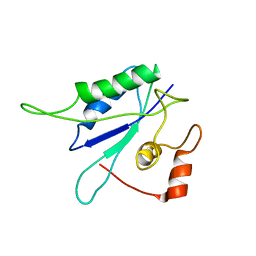 | |
3HCX
 
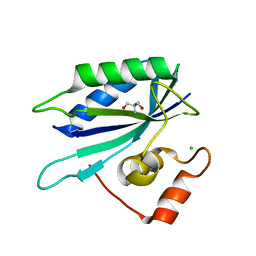 | | Crystal structure of E. coli HPPK(N10A) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, CHLORIDE ION | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-05-06 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Role of loop coupling in enzymatic catalysis and conformational dynamics
To be Published
|
|
3HD2
 
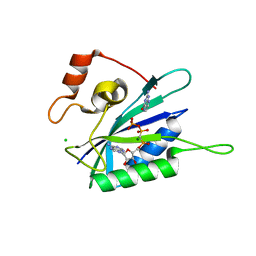 | | Crystal structure of E. coli HPPK(Q50A) in complex with MgAMPCPP and pterin | | Descriptor: | 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, CHLORIDE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-05-06 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Role of loop coupling in enzymatic catalysis and conformational dynamics
To be Published
|
|
5GZO
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.755 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
6V6R
 
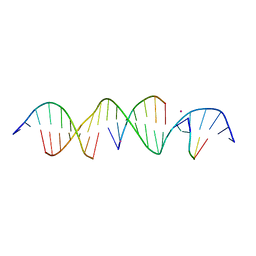 | | Crystal Structure of a Bromine Derivatized Self-Assembling DNA Crystal Scaffold with Rhombohedral Symmetry. | | Descriptor: | CACODYLATE ION, DNA (5'-D(*CP*AP*CP*TP*GP*AP*CP*TP*CP*AP*TP*GP*CP*TP*CP*AP*TP*CP*TP*GP*A)-3'), DNA (5'-D(*TP*GP*TP*CP*AP*GP*AP*TP*G)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2019-12-05 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Self-Assembled Rhombohedral DNA Crystal Scaffold with Tunable Cavity Sizes and High-Resolution Structural Detail.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
4BZB
 
 | | Crystal structure of the tetrameric dGTP-bound SAMHD1 mutant catalytic core | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DEOXYNUCLEOSIDE TRIPHOSPHATE TRIPHOSPHOHYDROLASE SAMHD1, MAGNESIUM ION | | Authors: | Ji, X, Yang, H, Wu, Y, Yan, J, Mehrens, J, DeLucia, M, Hao, C, Gronenborn, A.M, Skowronski, J, Ahn, J, Xiong, Y. | | Deposit date: | 2013-07-25 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Mechanism of Allosteric Activation of Samhd1 by Dgtp
Nat.Struct.Mol.Biol., 20, 2013
|
|
4BZC
 
 | | Crystal structure of the tetrameric dGTP-bound wild type SAMHD1 catalytic core | | Descriptor: | 2'-deoxyguanosine-5'-O-(1-thiotriphosphate), DEOXYNUCLEOSIDE TRIPHOSPHATE TRIPHOSPHOHYDROLASE SAMHD1, MAGNESIUM ION, ... | | Authors: | Ji, X, Yang, H, Wu, Y, Yan, J, Mehrens, J, DeLucia, M, Hao, C, Gronenborn, A.M, Skowronski, J, Ahn, J, Xiong, Y. | | Deposit date: | 2013-07-25 | | Release date: | 2013-10-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Mechanism of Allosteric Activation of Samhd1 by Dgtp
Nat.Struct.Mol.Biol., 20, 2013
|
|
5FNT
 
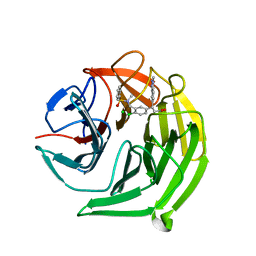 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-{4-Chloro-3-[(N-methylbenzenesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FZJ
 
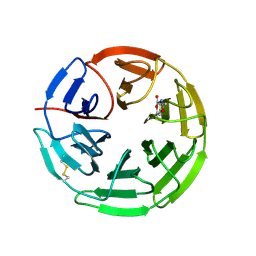 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | 2,6-DIMETHYL-4H-PYRANO[3,4-D][1,3]OXAZOL-4-ONE, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2016-03-14 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
1HKA
 
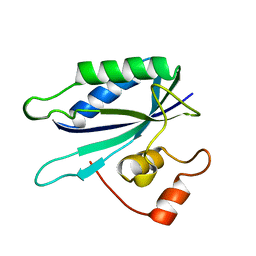 | | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE | | Descriptor: | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE | | Authors: | Xiao, B, Shi, G, Chen, X, Yan, H, Ji, X. | | Deposit date: | 1998-09-29 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, a potential target for the development of novel antimicrobial agents.
Structure Fold.Des., 7, 1999
|
|
8SSX
 
 | | Crystal structure of Bacillus anthracis dihydrofolate reductase at 1.65-A resolution | | Descriptor: | 1,2-ETHANEDIOL, Dihydrofolate reductase, SULFATE ION | | Authors: | Shaw, G.X, Li, Y, Wu, Y, Yan, H, Ji, X. | | Deposit date: | 2023-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Bacillus anthracis dihydrofolate reductase at 1.65-A resolution
To be published
|
|
8SU7
 
 | | Crystal structure of Bacillus anthracis dihydroneopterin aldolase | | Descriptor: | 1,2-ETHANEDIOL, 7,8-dihydroneopterin aldolase | | Authors: | Shaw, G.X, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2023-05-11 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Bacillus anthracis dihydroneopterin aldolase
To be published
|
|
5GZN
 
 | | Structure of neutralizing antibody bound to Zika envelope protein | | Descriptor: | Antibody Heavy chain, Antibody light chain, Genome polyprotein | | Authors: | Wang, Q, Yang, H, Liu, X, Dai, L, Ma, T, Qi, J, Wong, G, Peng, R, Liu, S, Li, J, Li, S, Song, J, Liu, J, He, J, Yuan, H, Xiong, Y, Liao, Y, Li, J, Yang, J, Tong, Z, Griffin, B, Bi, Y, Liang, M, Xu, X, Cheng, G, Wang, P, Qiu, X, Kobinger, G, Shi, Y, Yan, J, Gao, G.F. | | Deposit date: | 2016-09-29 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular determinants of human neutralizing antibodies isolated from a patient infected with Zika virus
Sci Transl Med, 8, 2016
|
|
6XDX
 
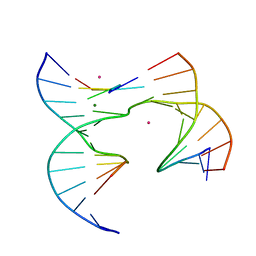 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J6 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*AP*TP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-11 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6X8B
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 duplex version) containing the J6 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*AP*AP*GP*AP*CP*TP*TP*CP*AP*CP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*A)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.899 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6X8C
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J1 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*TP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-01 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
