4Z5C
 
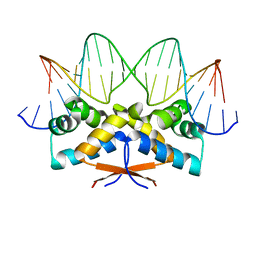 | | HipB-O3 21mer complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*TP*TP*TP*AP*TP*CP*CP*CP*GP*TP*AP*GP*AP*GP*CP*GP*GP*AP*TP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*TP*AP*CP*GP*GP*GP*AP*TP*A)-3') | | Authors: | Min, J, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2015-04-02 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism on hipBA gene regulation.
To be published
|
|
5EUN
 
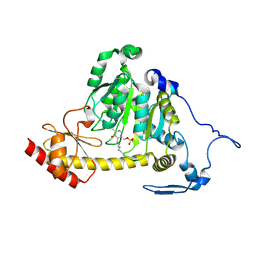 | | The Crystal Structure of Human Kynurenine Aminotransferase II, PLP-bound form, at 1.83 A | | Descriptor: | Kynurenine/alpha-aminoadipate aminotransferase, mitochondrial | | Authors: | Nematollahi, A, Sun, G, Kwan, A, Jeffries, C.M, Harrop, S.J, Hanrahan, J.R, Nadvi, N.A, Church, W.B. | | Deposit date: | 2015-11-18 | | Release date: | 2015-12-16 | | Last modified: | 2016-08-10 | | Method: | X-RAY DIFFRACTION (1.825 Å) | | Cite: | Structure of the PLP-Form of the Human Kynurenine Aminotransferase II in a Novel Spacegroup at 1.83 angstrom Resolution.
Int J Mol Sci, 17, 2016
|
|
4Z5D
 
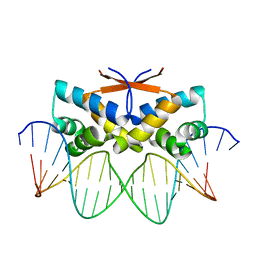 | | HipB-O4 21mer complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*TP*TP*TP*AP*TP*CP*CP*GP*CP*GP*AP*TP*CP*GP*CP*GP*GP*AP*TP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*TP*CP*CP*GP*CP*GP*AP*TP*CP*GP*CP*GP*GP*AP*TP*AP*A)-3') | | Authors: | Min, J, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2015-04-02 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular mechanism on hipBA gene regulation.
To be published
|
|
4Z59
 
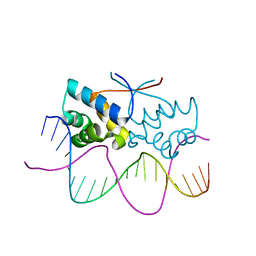 | | HipB-O4 20mer complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*TP*TP*AP*TP*CP*CP*GP*CP*GP*AP*TP*CP*GP*CP*GP*GP*AP*TP*AP*A)-3') | | Authors: | Min, J, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2015-04-02 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular mechanism on hipBA gene regulation.
To be published
|
|
4Z5H
 
 | | HipB(S29A)-O2 20mer complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*TP*TP*AP*TP*CP*CP*TP*CP*AP*CP*TP*AP*AP*AP*GP*GP*AP*TP*AP*A)-3') | | Authors: | Min, J, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2015-04-02 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism on hipBA gene regulation.
To be published
|
|
6VL8
 
 | | Anti-PEG antibody 6-3 Fab fragment in complex with PEG | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, 6-3 Fab heavy chain, ... | | Authors: | Nicely, N.I, Huckaby, J.T, Lai, S.K, Jacobs, T.M. | | Deposit date: | 2020-01-23 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of an anti-PEG antibody reveals an open ring that captures highly flexible PEG polymers
Commun Chem, 3, 2020
|
|
7LSY
 
 | | NHEJ Short-range synaptic complex | | Descriptor: | DNA (26-MER), DNA (5'-D(P*CP*AP*AP*TP*GP*AP*AP*AP*CP*GP*GP*AP*AP*CP*AP*GP*TP*CP*AP*G)-3'), DNA (5'-D(P*GP*TP*TP*CP*TP*TP*AP*GP*TP*AP*TP*AP*TP*A)-3'), ... | | Authors: | He, Y, Chen, S. | | Deposit date: | 2021-02-18 | | Release date: | 2021-04-14 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structural basis of long-range to short-range synaptic transition in NHEJ.
Nature, 593, 2021
|
|
6VL9
 
 | | Anti-PEG antibody 6-3 Fab fragment in complex with PEG | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39,42,45,48,51,54,57-nonadecaoxanonapentacontane-1,59-diol, 6-3 Fab heavy chain, 6-3 Fab light chain | | Authors: | Nicely, N.I, Huckaby, J.T, Lai, S.K, Jacobs, T.M. | | Deposit date: | 2020-01-23 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structure of an anti-PEG antibody reveals an open ring that captures highly flexible PEG polymers
Commun Chem, 3, 2020
|
|
4BWH
 
 | |
5MY7
 
 | | Adhesin Complex Protein from Neisseria meningitidis | | Descriptor: | Adhesin, DI(HYDROXYETHYL)ETHER, SODIUM ION | | Authors: | Derrick, J.P, Awanye, A. | | Deposit date: | 2017-01-25 | | Release date: | 2017-06-21 | | Last modified: | 2017-07-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Neisseria Adhesin Complex Protein (ACP) and its role as a novel lysozyme inhibitor.
PLoS Pathog., 13, 2017
|
|
1YQ3
 
 | | Avian respiratory complex ii with oxaloacetate and ubiquinone | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Coenzyme Q10, (2Z,6E,10Z,14E,18E,22E,26Z)-isomer, ... | | Authors: | Huang, L, Cobessi, D, Tung, E.Y, Berry, E.A. | | Deposit date: | 2005-02-01 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 3-Nitropropionic Acid Is a Suicide Inhibitor of Mitochondrial Respiration That, upon Oxidation by Complex II, Forms a Covalent Adduct with a Catalytic Base Arginine in the Active Site of the Enzyme
J.Biol.Chem., 281, 2006
|
|
5H1D
 
 | | Crystal structure of C-terminal of RhoGDI2 | | Descriptor: | Rho GDP-dissociation inhibitor 2 | | Authors: | Liu, J. | | Deposit date: | 2016-10-08 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.494 Å) | | Cite: | NMR characterization of weak interactions between RhoGDI2 and fragment screening hits.
Biochim. Biophys. Acta, 1861, 2017
|
|
6K0L
 
 | | The crystal structure of simian CD163 SRCR5 | | Descriptor: | Scavenger receptor cysteine-rich type 1 protein M130 | | Authors: | Ma, H, Li, R, Jiang, L, Qiao, S, Zhang, G. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural comparison of CD163 SRCR5 from different species sheds some light on its involvement in porcine reproductive and respiratory syndrome virus-2 infection in vitro.
Vet Res, 52, 2021
|
|
6K0O
 
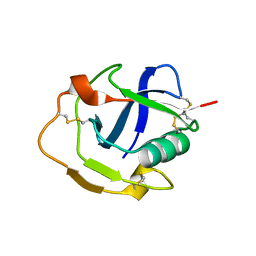 | | The crystal structure of human CD163-like homolog SRCR8 | | Descriptor: | Scavenger receptor cysteine-rich type 1 protein M160 | | Authors: | Ma, H, Li, R, Jiang, L, Qiao, S, Zhang, G. | | Deposit date: | 2019-05-07 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural comparison of CD163 SRCR5 from different species sheds some light on its involvement in porcine reproductive and respiratory syndrome virus-2 infection in vitro.
Vet Res, 52, 2021
|
|
2FBW
 
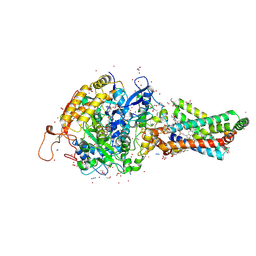 | | Avian respiratory complex II with carboxin bound | | Descriptor: | (~{Z})-2-oxidanylbut-2-enedioic acid, 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, AZIDE ION, ... | | Authors: | Huang, L.S, Sun, G, Cobessi, D, Wang, A.C, Shen, J.T, Tung, E.Y, Anderson, V.E, Berry, E.A. | | Deposit date: | 2005-12-10 | | Release date: | 2005-12-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | 3-nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme.
J.Biol.Chem., 281, 2006
|
|
6MOM
 
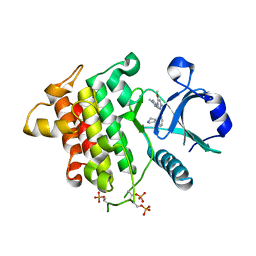 | | Crystal structure of human Interleukin-1 receptor associated Kinase 4 (IRAK 4, CID 100300) in complex with compound NCC00371481 (BSI 107591) | | Descriptor: | 1,2-ETHANEDIOL, 6-[7-methoxy-6-(1-methyl-1H-pyrazol-4-yl)imidazo[1,2-a]pyridin-3-yl]-N-[(3R)-pyrrolidin-3-yl]pyridin-2-amine, Interleukin-1 receptor-associated kinase 4 | | Authors: | Abendroth, J, Mayclin, S.J, Lorimer, D.D, Starczynowski, D, Hoyt, S, Tawa, G, Thomas, C. | | Deposit date: | 2018-10-04 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Overcoming adaptive therapy resistance in AML by targeting immune response pathways.
Sci Transl Med, 11, 2019
|
|
2H89
 
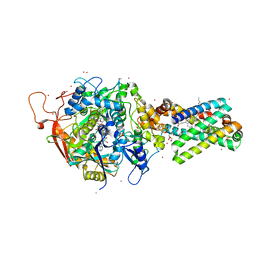 | | Avian Respiratory Complex II with Malonate Bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Huang, L.S, Shen, J.T, Wang, A.C, Berry, E.A. | | Deposit date: | 2006-06-06 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the
Biochim.Biophys.Acta, 1757
|
|
2H88
 
 | | Avian Mitochondrial Respiratory Complex II at 1.8 Angstrom Resolution | | Descriptor: | AZIDE ION, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Huang, L.S, Shen, J.T, Wang, A.C, Berry, E.A. | | Deposit date: | 2006-06-06 | | Release date: | 2006-06-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystallographic studies of the binding of ligands to the dicarboxylate site of Complex II, and the identity of the ligand in the
Biochim.Biophys.Acta, 1757
|
|
2LXY
 
 | | NMR structure of 2-MERCAPTOPHENOL-ALPHA3C | | Descriptor: | 2-MERCAPTOPHENOL, 2-mercaptophenol-alpha3C | | Authors: | Tommos, C, Valentine, K.G, Martinez-Rivera, M.C, Liang, L, Moorman, V.R. | | Deposit date: | 2012-09-06 | | Release date: | 2013-02-27 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Reversible phenol oxidation and reduction in the structurally well-defined 2-Mercaptophenol-alpha(3)C protein.
Biochemistry, 52, 2013
|
|
5NBH
 
 | |
5NC1
 
 | |
4IAN
 
 | | Crystal Structure of apo Human PRPF4B kinase domain | | Descriptor: | SULFATE ION, Serine/threonine-protein kinase PRP4 homolog | | Authors: | Mechin, I, Haas, K, Chen, X, Zhang, Y, McLean, L. | | Deposit date: | 2012-12-06 | | Release date: | 2013-08-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Evaluation of Cancer Dependence and Druggability of PRP4 Kinase Using Cellular, Biochemical, and Structural Approaches.
J.Biol.Chem., 288, 2013
|
|
1WUP
 
 | | Crystal structure of metallo-beta-lactamase IMP-1 mutant (D81E) | | Descriptor: | ACETIC ACID, Beta-lactamase IMP-1, ZINC ION | | Authors: | Yamaguchi, Y, Yamagata, Y, Goto, M. | | Deposit date: | 2004-12-08 | | Release date: | 2005-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Probing the role of Asp-120(81) of metallo-beta-lactamase (IMP-1) by site-directed mutagenesis, kinetic studies, and X-ray crystallography.
J.Biol.Chem., 280, 2005
|
|
7BB3
 
 | | Structure of S. pombe YG-box oligomer | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Survival motor neuron-like protein 1,Survival motor neuron-like protein 1 | | Authors: | Veepaschit, J, Grimm, C, Fischer, U. | | Deposit date: | 2020-12-16 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.158 Å) | | Cite: | Identification and structural analysis of the Schizosaccharomyces pombe SMN complex.
Nucleic Acids Res., 49, 2021
|
|
1WUO
 
 | | Crystal structure of metallo-beta-lactamase IMP-1 mutant (D81A) | | Descriptor: | ACETIC ACID, Beta-lactamase IMP-1, ZINC ION | | Authors: | Yamaguchi, Y, Yamagata, Y, Goto, M. | | Deposit date: | 2004-12-08 | | Release date: | 2005-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Probing the role of Asp-120(81) of metallo-beta-lactamase (IMP-1) by site-directed mutagenesis, kinetic studies, and X-ray crystallography.
J.Biol.Chem., 280, 2005
|
|
