9FQH
 
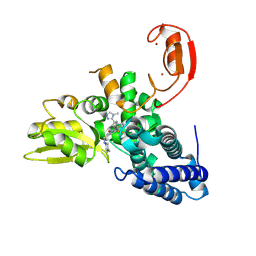 | | E3 ligase Cbl-b in complex with a triazolone core inhibitor (compound 1) | | Descriptor: | 8-[3-[3-methyl-1-(4-methyl-1,2,4-triazol-3-yl)cyclobutyl]phenyl]-3-[[(3~{S})-3-methylpiperidin-1-yl]methyl]-5-(trifluoromethyl)-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2024-06-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Accelerated Discovery of Carbamate Cbl-b Inhibitors Using Generative AI Models and Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
9FQJ
 
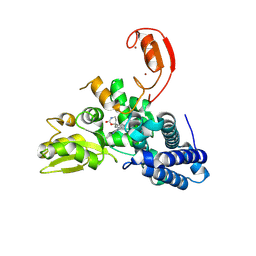 | | E3 ligase Cbl-b in complex with a carbamate scaffold inhibitor (compound 12) | | Descriptor: | 2-cyclopropyl-6-methyl-~{N}-[3-[(6~{S})-6-methyl-2-oxidanylidene-1,3-oxazinan-6-yl]phenyl]pyrimidine-4-carboxamide, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2024-06-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.563 Å) | | Cite: | Accelerated Discovery of Carbamate Cbl-b Inhibitors Using Generative AI Models and Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
9FQI
 
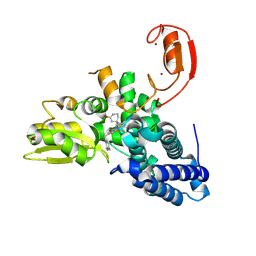 | | E3 ligase Cbl-b in complex with a lactam scaffold inhibitor (compound 7) | | Descriptor: | 8-[3-[(4~{R})-4-methyl-2-oxidanylidene-piperidin-4-yl]phenyl]-3-[[(3~{S})-3-methylpiperidin-1-yl]methyl]-5-(trifluoromethyl)-1$l^{4},7,8-triazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, E3 ubiquitin-protein ligase CBL-B, SODIUM ION, ... | | Authors: | Schimpl, M. | | Deposit date: | 2024-06-17 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Accelerated Discovery of Carbamate Cbl-b Inhibitors Using Generative AI Models and Structure-Based Drug Design.
J.Med.Chem., 67, 2024
|
|
8J7N
 
 | |
8J7P
 
 | |
3LHG
 
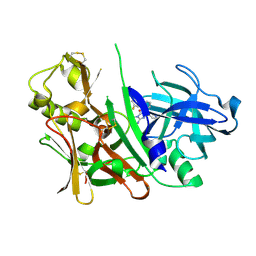 | | Bace1 in complex with the aminohydantoin Compound 4g | | Descriptor: | (5S)-2-amino-5-(2',5'-difluorobiphenyl-3-yl)-3-methyl-5-pyridin-4-yl-3,5-dihydro-4H-imidazol-4-one, Beta-secretase 1 | | Authors: | Olland, A.M. | | Deposit date: | 2010-01-22 | | Release date: | 2010-04-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyridinyl aminohydantoins as small molecule BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3MTN
 
 | | Usp21 in complex with a ubiquitin-based, USP21-specific inhibitor | | Descriptor: | CHLORIDE ION, GLYCEROL, UBIQUITIN VARIANT UBV.21.4, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Ernst, A, Sidhu, S, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-30 | | Release date: | 2010-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
3N3K
 
 | | The catalytic domain of USP8 in complex with a USP8 specific inhibitor | | Descriptor: | Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 8, ZINC ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Allali-Hassani, A, Lam, R, Ernst, A, Sidhu, S, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-20 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
5E1R
 
 | |
3GFV
 
 | | Crystal Structure of Petrobactin-binding Protein YclQ from Bacillu subtilis | | Descriptor: | ASPARAGINE, PHOSPHATE ION, Uncharacterized ABC transporter solute-binding protein yclQ | | Authors: | Kim, Y, Maltseva, N, Zawadzka, A.M, Raymond, K.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-27 | | Release date: | 2009-05-12 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization of a Bacillus subtilis transporter for petrobactin, an anthrax stealth siderophore.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8FUR
 
 | | Crystal structure of human IDO1 with compound 11 | | Descriptor: | Indoleamine 2,3-dioxygenase 1, N-(4-methylphenyl)-N'-[(1P,2'P)-4-propoxy-5-propyl-2'-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-yl]urea | | Authors: | Critton, D.A, Lewis, H.A. | | Deposit date: | 2023-01-18 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (2.285 Å) | | Cite: | Synthesis and biological evaluation of biaryl alkyl ethers as inhibitors of IDO1.
Bioorg.Med.Chem.Lett., 88, 2023
|
|
3I3T
 
 | | Crystal structure of covalent ubiquitin-USP21 complex | | Descriptor: | ETHANAMINE, Ubiquitin, Ubiquitin carboxyl-terminal hydrolase 21, ... | | Authors: | Neculai, D, Avvakumov, G.V, Walker, J.R, Xue, S, Butler-Cole, C, Weigelt, J, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
3V5Q
 
 | | Discovery of a selective TRK Inhibitor with efficacy in rodent cancer tumor models | | Descriptor: | 1-(3-{[(3Z)-2-oxo-3-(1H-pyrrol-2-ylmethylidene)-2,3-dihydro-1H-indol-6-yl]amino}phenyl)-3-[3-(trifluoromethyl)phenyl]urea, CHLORIDE ION, NT-3 growth factor receptor | | Authors: | Kreusch, A. | | Deposit date: | 2011-12-16 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2001 Å) | | Cite: | Discovery of GNF-5837, a Selective TRK Inhibitor with Efficacy in Rodent Cancer Tumor Models.
ACS Med Chem Lett, 3, 2012
|
|
7SN0
 
 | | Crystal structure of spike protein receptor binding domain of escape mutant SARS-CoV-2 from immunocompromised patient (d146*) in complex with human receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Abraham, J, Clark, S. | | Deposit date: | 2021-10-27 | | Release date: | 2021-12-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
7SN1
 
 | |
7SN3
 
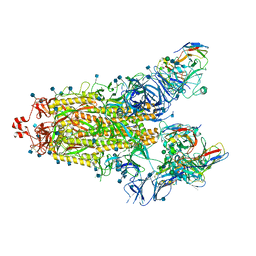 | | Structure of human SARS-CoV-2 spike glycoprotein trimer bound by neutralizing antibody C1C-A3 Fab (variable region) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Abraham, J, Shankar, S. | | Deposit date: | 2021-10-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
7SN2
 
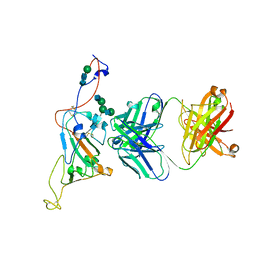 | | Structure of human SARS-CoV-2 neutralizing antibody C1C-A3 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Abraham, J, Yang, P, Shankar, S. | | Deposit date: | 2021-10-27 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for continued antibody evasion by the SARS-CoV-2 receptor binding domain.
Science, 375, 2022
|
|
4I6L
 
 | |
3V6E
 
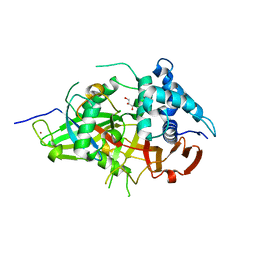 | | Crystal Structure of USP2 and a mutant form of Ubiquitin | | Descriptor: | CHLORIDE ION, GLYCEROL, Ubiquitin, ... | | Authors: | Neculai, M, Ernst, A, Sidhu, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-19 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
8WCP
 
 | |
7ZQY
 
 | | Chaetomium thermophilum Rad50 Zn hook | | Descriptor: | DH domain-containing protein, ZINC ION | | Authors: | Lammens, K, Rotheneder, M, Stakyte, K. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
7ZR1
 
 | | Chaetomium thermophilum Mre11-Rad50-Nbs1 complex bound to ATPyS (composite structure) | | Descriptor: | DH domain-containing protein, Double-strand break repair protein, FHA domain-containing protein, ... | | Authors: | Bartho, J.D, Rotheneder, M, Stakyte, K, Lammens, K, Hopfner, K.P. | | Deposit date: | 2022-05-03 | | Release date: | 2023-01-11 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the Mre11-Rad50-Nbs1 complex reveals the molecular mechanism of scaffolding functions.
Mol.Cell, 83, 2023
|
|
7L4S
 
 | |
3NWW
 
 | | P38 Alpha kinase complexed with a 2-aminothiazol-5-yl-pyrimidine based inhibitor | | Descriptor: | 1-[2-(2-{[2-(dimethylamino)ethyl]amino}-6-{2-[(1-methylethyl)amino]-1,3-thiazol-5-yl}pyrimidin-4-yl)benzyl]-3-ethylurea, Mitogen-activated protein kinase 14 | | Authors: | Sack, J.S. | | Deposit date: | 2010-07-12 | | Release date: | 2010-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Utilization of a nitrogen-sulfur nonbonding interaction in the design of new 2-aminothiazol-5-yl-pyrimidines as p38alpha MAP kinase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
7LT6
 
 | | Structure of Partial Beta-Hairpin LIR from FNIP2 Bound to GABARAP | | Descriptor: | Folliculin-interacting protein 2,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Appleton, B.A. | | Deposit date: | 2021-02-18 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | GABARAP sequesters the FLCN-FNIP tumor suppressor complex to couple autophagy with lysosomal biogenesis.
Sci Adv, 7, 2021
|
|
