8PGL
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3027 | | Descriptor: | 7-[(1~{S})-1-[5-(carbamimidamidomethyl)-2-oxidanylidene-1,3-oxazolidin-3-yl]ethyl]-3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, DIMETHYL SULFOXIDE, ... | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3027
To Be Published
|
|
8PG5
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 2731 | | Descriptor: | 3-[3-fluoranyl-4-(methylsulfonylmethyl)phenyl]-7-[(1~{S})-1-(piperidin-4-ylmethoxy)ethyl]-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 2731
To Be Published
|
|
8PGM
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3062 | | Descriptor: | 3-[4-[(azanyl-methyl-oxidanyl-$l^{4}-sulfanyl)methyl]-3-fluoranyl-phenyl]-7-propan-2-yl-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, DIMETHYL SULFOXIDE, ... | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3062
To Be Published
|
|
8PGZ
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3528 | | Descriptor: | 7-[(1~{S})-1-[2-(aminomethyl)-6-oxidanylidene-5-oxa-7-azaspiro[3.4]octan-7-yl]ethyl]-3-(6-oxidanylidene-1~{H}-pyridazin-4-yl)-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3528
To Be Published
|
|
8PA0
 
 | | cvHsp (HspB7) C131S alpha-crystallin domain - filamin C (FLNC) domain 24 complex | | Descriptor: | Filamin-C, Heat shock protein beta-7 | | Authors: | Wang, Z, Benesch, J.L.P, Allison, T.M, Song, H, McDonough, M.A, Brem, J, Rabe, P. | | Deposit date: | 2023-06-06 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | cvHsp (HspB7) C131S alpha-crystallin domain - filamin C (FLNC) domain 24 complex
To Be Published
|
|
7DML
 
 | |
7E9A
 
 | | Crystal structure of KPC-2 in complex with (S)-2-(1-hydroxy-1,3-dihydrobenzo[c][1,2]oxaborol-3-yl)acrylic acid (4a-(S)) | | Descriptor: | 2-[(3S)-1-oxidanyl-3H-2,1-benzoxaborol-3-yl]prop-2-enoic acid, ACETIC ACID, Beta-lactamase, ... | | Authors: | Li, G.-B, Yan, Y.-H. | | Deposit date: | 2021-03-03 | | Release date: | 2021-08-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design and enantioselective synthesis of 3-( alpha-acrylic acid) benzoxaboroles to combat carbapenemase resistance.
Chem.Commun.(Camb.), 57, 2021
|
|
4KQQ
 
 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | Descriptor: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4KQO
 
 | | Crystal structure of penicillin-binding protein 3 from pseudomonas aeruginosa in complex with piperacillin | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
4KQR
 
 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | Descriptor: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
6QWB
 
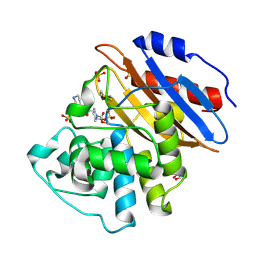 | | Crystal structure of KPC-4 complexed with relebactam (16 hour soak) | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, (2~{S})-5-azanylidene-2-(piperidin-4-ylcarbamoyl)piperidine-1-carboxylic acid, Beta-lactamase, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-03-05 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Molecular Basis of Class A beta-Lactamase Inhibition by Relebactam.
Antimicrob.Agents Chemother., 63, 2019
|
|
6QWD
 
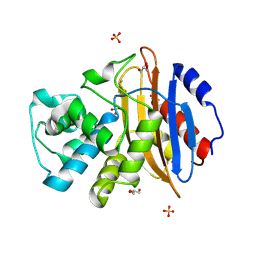 | | Crystal structure of KPC-3 | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-03-05 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Molecular Basis of Class A beta-Lactamase Inhibition by Relebactam.
Antimicrob.Agents Chemother., 63, 2019
|
|
6QWA
 
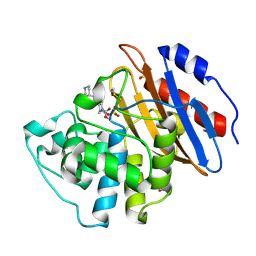 | | Crystal structure of KPC-3 complexed with relebactam (16 hour soak) | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, (2~{S})-5-azanylidene-2-(piperidin-4-ylcarbamoyl)piperidine-1-carboxylic acid, Beta-lactamase, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-03-05 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Molecular Basis of Class A beta-Lactamase Inhibition by Relebactam.
Antimicrob.Agents Chemother., 63, 2019
|
|
6QW8
 
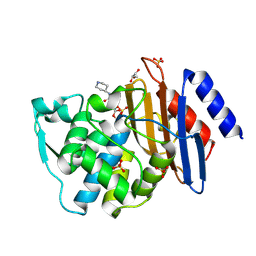 | | Crystal structure of CTX-M-15 complexed with relebactam (16 hour soak) | | Descriptor: | (2S,5R)-1-formyl-N-(piperidin-4-yl)-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, GLYCEROL, ... | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-03-05 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular Basis of Class A beta-Lactamase Inhibition by Relebactam.
Antimicrob.Agents Chemother., 63, 2019
|
|
6QWE
 
 | | Crystal structure of KPC-4 | | Descriptor: | Beta-lactamase, GLYCEROL, SULFATE ION | | Authors: | Tooke, C.L, Hinchliffe, P, Spencer, J. | | Deposit date: | 2019-03-05 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Basis of Class A beta-Lactamase Inhibition by Relebactam.
Antimicrob.Agents Chemother., 63, 2019
|
|
6Y0P
 
 | | isopenicillin N synthase in complex with IPN and Fe using FT-SSX methods | | Descriptor: | FE (III) ION, ISOPENICILLIN N, Isopenicillin N synthase, ... | | Authors: | Rabe, P, Beale, J.H, Lang, P.A, Dirr, A.S, Leissing, T.M, Butryn, A, Aller, P, Kamps, J.J.A.G, Axford, D, McDonough, M.A, Orville, A.M, Owen, R, Schofield, C.J. | | Deposit date: | 2020-02-10 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray free-electron laser studies reveal correlated motion during isopenicillin N synthase catalysis.
Sci Adv, 7, 2021
|
|
5IQK
 
 | | Rm3 metallo-beta-lactamase | | Descriptor: | ZINC ION, beta-lactamase Rm3 | | Authors: | Salimraj, R, Spencer, J. | | Deposit date: | 2016-03-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biochemical Characterization of Rm3, a Subclass B3 Metallo-beta-Lactamase Identified from a Functional Metagenomic Study.
Antimicrob.Agents Chemother., 60, 2016
|
|
8C8S
 
 | | Crystal structure of human DNA cross-link repair 1A in complex with hydroxamic acid inhibitor (compound 21). | | Descriptor: | (2~{R})-3-[6-chloranyl-2-(prop-2-enylamino)quinazolin-4-yl]-2-methyl-~{N}-oxidanyl-propanamide, DNA cross-link repair 1A protein, ZINC ION | | Authors: | Yosaatmadja, Y, Newman, J.A, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-01-20 | | Release date: | 2024-01-31 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cell-active small molecule inhibitors validate the SNM1A DNA repair nuclease as a cancer target.
Chem Sci, 15, 2024
|
|
8C8B
 
 | | Crystal structure of human DNA cross-link repair 1A in complex with hydroxamic acid inhibitor (compound 48). | | Descriptor: | 4-[[(2~{S})-1-(oxidanylamino)-1-oxidanylidene-propan-2-yl]amino]-~{N}-prop-2-enyl-quinazoline-2-carboxamide, DNA cross-link repair 1A protein, ZINC ION | | Authors: | Yosaatmadja, Y, Newman, J.A, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-31 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Cell-active small molecule inhibitors validate the SNM1A DNA repair nuclease as a cancer target.
Chem Sci, 15, 2024
|
|
8C8D
 
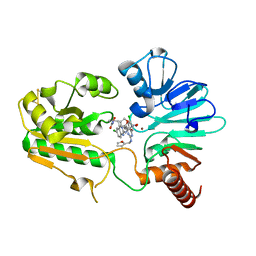 | | Crystal structure of human DNA cross-link repair 1A in complex with hydroxamic acid inhibitor (compound 44). | | Descriptor: | (2~{R})-3-[6-chloranyl-2-(furan-2-ylmethylamino)quinazolin-4-yl]-2-methyl-~{N}-oxidanyl-propanamide, DNA cross-link repair 1A protein, ZINC ION | | Authors: | Yosaatmadja, Y, Newman, J.A, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Cell-active small molecule inhibitors validate the SNM1A DNA repair nuclease as a cancer target.
Chem Sci, 15, 2024
|
|
8CEW
 
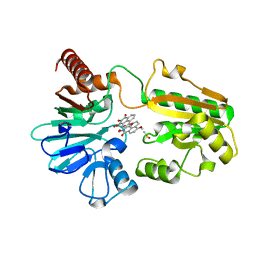 | | Crystal structure of human DNA cross-link repair 1A in complex with N-hydroxyimide inhibitor H1 | | Descriptor: | 6-methoxy-2-oxidanyl-benzo[de]isoquinoline-1,3-dione, DIMETHYL SULFOXIDE, DNA cross-link repair 1A protein, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Cell-active small molecule inhibitors validate the SNM1A DNA repair nuclease as a cancer target.
Chem Sci, 15, 2024
|
|
8CF0
 
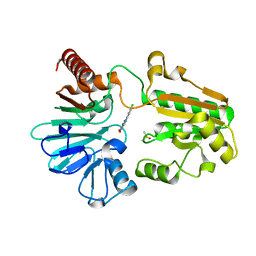 | | Crystal structure of human DNA cross-link repair 1A in complex with quinoxalinedione inhibitor H2 | | Descriptor: | 9-chloranyl-1,4-dihydropyrazino[2,3-c]quinoline-2,3-dione, DIMETHYL SULFOXIDE, DNA cross-link repair 1A protein, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Cell-active small molecule inhibitors validate the SNM1A DNA repair nuclease as a cancer target.
Chem Sci, 15, 2024
|
|
8CG9
 
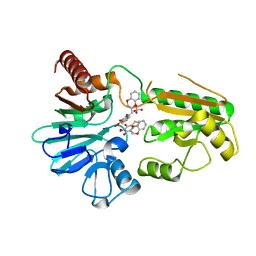 | | Crystal structure of human DNA cross-link repair 1A in complex with a cyclic N-hydroxyurea inhibitor | | Descriptor: | 1-[(2S)-2,3-dihydro-1,4-benzodioxin-2-ylmethyl]-3-hydroxythieno[3,2-d]pyrimidine-2,4(1H,3H)-dione, 1-[[(3~{R})-2,3-dihydro-1,4-benzodioxin-3-yl]methyl]-3-oxidanyl-thieno[3,2-d]pyrimidine-2,4-dione, DNA cross-link repair 1A protein, ... | | Authors: | Newman, J.A, Yosaatmadja, Y, Baddock, H.T, Bielinski, M, von Delft, F, Bountra, C, McHugh, P.J, Schofield, C.J, Gileadi, O. | | Deposit date: | 2023-02-03 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Cell-active small molecule inhibitors validate the SNM1A DNA repair nuclease as a cancer target.
Chem Sci, 15, 2024
|
|
5N5H
 
 | | Crystal structure of metallo-beta-lactamase VIM-1 in complex with ML302F inhibitor | | Descriptor: | (2Z)-2-sulfanyl-3-(2,3,6-trichlorophenyl)prop-2-enoic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Salimraj, R, Hinchliffe, P, Spencer, J. | | Deposit date: | 2017-02-14 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structures of VIM-1 complexes explain active site heterogeneity in VIM-class metallo-beta-lactamases.
FEBS J., 286, 2019
|
|
5N5I
 
 | | Crystal Structure of VIM-1 metallo-beta-lactamase in complex with hydrolysed meropenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Salimraj, R, Hinchliffe, P, Spencer, J. | | Deposit date: | 2017-02-14 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of VIM-1 complexes explain active site heterogeneity in VIM-class metallo-beta-lactamases.
FEBS J., 286, 2019
|
|
