2P4S
 
 | | Structure of Purine Nucleoside Phosphorylase from Anopheles gambiae in complex with DADMe-ImmH | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Rinaldo-Matthis, A, Almo, S.C, Schramm, V.L. | | Deposit date: | 2007-03-13 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anopheles gambiae purine nucleoside phosphorylase: catalysis, structure, and inhibition.
Biochemistry, 46, 2007
|
|
7M33
 
 | |
3ZNZ
 
 | | Crystal structure of OTULIN OTU domain (C129A) in complex with Met1- di ubiquitin | | Descriptor: | POLYUBIQUITIN-C, PROTEIN FAM105B, SULFATE ION | | Authors: | Keusekotten, K, Elliott, P.R, Glockner, L, Kulathu, Y, Wauer, T, Krappmann, D, Hofmann, K, Komander, D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Otulin Antagonizes Lubac Signaling by Specifically Hydrolyzing met1-Linked Polyubiquitin.
Cell(Cambridge,Mass.), 153, 2013
|
|
3K35
 
 | | Crystal Structure of Human SIRT6 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent deacetylase sirtuin-6, SULFATE ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
4JAI
 
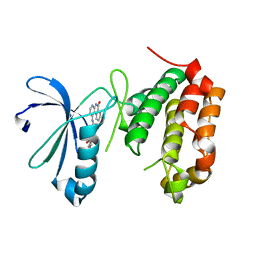 | | Crystal Structure of Aurora Kinase A in complex with N-{4-[(6-oxo-5,6-dihydrobenzo[c][1,8]naphthyridin-1-yl)amino]phenyl}benzamide | | Descriptor: | Aurora kinase A, N-{4-[(6-oxo-5,6-dihydrobenzo[c][1,8]naphthyridin-1-yl)amino]phenyl}benzamide | | Authors: | Jiang, X, Josephson, K, Huck, B, Goutopoulos, A, Karra, S. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | SAR and evaluation of novel 5H-benzo[c][1,8]naphthyridin-6-one analogs as Aurora kinase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4J8V
 
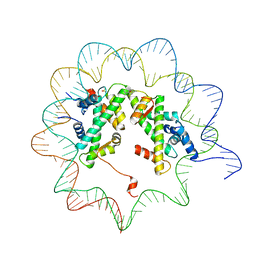 | |
4J8U
 
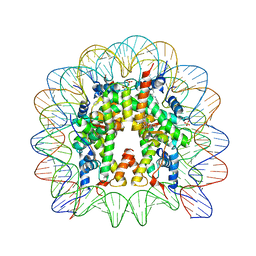 | |
4CPK
 
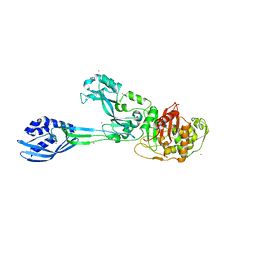 | | Crystal structure of PBP2a double clinical mutant N146K-E150K from MRSA | | Descriptor: | CADMIUM ION, CHLORIDE ION, Penicillin binding protein 2 prime, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Hermoso, J.A. | | Deposit date: | 2014-02-07 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics.
J.Am.Chem.Soc., 136, 2014
|
|
4BL3
 
 | | Crystal structure of PBP2a clinical mutant N146K from MRSA | | Descriptor: | CADMIUM ION, CHLORIDE ION, PENICILLIN BINDING PROTEIN 2 PRIME, ... | | Authors: | Otero, L.H, Rojas-Altuve, A, Carrasco-Lopez, C, Hermoso, J.A. | | Deposit date: | 2013-04-30 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Disruption of Allosteric Response as an Unprecedented Mechanism of Resistance to Antibiotics.
J.Am.Chem.Soc., 136, 2014
|
|
3VOZ
 
 | | Crystal structure of human glutaminase in complex with BPTES | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide), ... | | Authors: | Thangavelu, K, Sivaraman, J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4JAJ
 
 | | Crystal Structure of Aurora Kinase A in complex with BENZO[C][1,8]NAPHTHYRIDIN-6(5H)-ONE | | Descriptor: | Aurora kinase A, benzo[c][1,8]naphthyridin-6(5H)-one | | Authors: | Jiang, X, Josephson, K, Huck, B, Goutopoulos, A, Karra, S. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SAR and evaluation of novel 5H-benzo[c][1,8]naphthyridin-6-one analogs as Aurora kinase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
2OOW
 
 | | MIF Bound to a Fluorinated OXIM Derivative | | Descriptor: | 3-FLUORO-4-HYDROXYBENZALDEHYDE O-(CYCLOHEXYLCARBONYL)OXIME, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Crichlow, G.V, Al-Abed, Y, Lolis, E. | | Deposit date: | 2007-01-26 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Alternative chemical modifications reverse the binding orientation of a pharmacophore scaffold in the active site of macrophage migration inhibitory factor.
J.Biol.Chem., 282, 2007
|
|
3VP2
 
 | | Crystal structure of human glutaminase in complex with inhibitor 2 | | Descriptor: | 5,5'-(sulfanediyldiethane-2,1-diyl)bis(1,3,4-thiadiazol-2-amine), Glutaminase kidney isoform, mitochondrial, ... | | Authors: | Thangavelu, K, Sivaraman, J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VP0
 
 | | Crystal structure of human glutaminase in complex with L-glutamine | | Descriptor: | GLUTAMINE, Glutaminase kidney isoform, mitochondrial, ... | | Authors: | Thangavelu, K, Sivaraman, J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VOY
 
 | | Crystal structure of human glutaminase in apo form | | Descriptor: | Glutaminase kidney isoform, mitochondrial, SULFATE ION | | Authors: | Thangavelu, K, Sivaraman, J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZNX
 
 | | Crystal structure of the OTU domain of OTULIN D336A mutant | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Keusekotten, K, Elliott, P.R, Glockner, L, Kulathu, Y, Wauer, T, Krappmann, D, Hofmann, K, Komander, D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Otulin Antagonizes Lubac Signaling by Specifically Hydrolyzing met1-Linked Polyubiquitin.
Cell(Cambridge,Mass.), 153, 2013
|
|
4J8X
 
 | |
3VP3
 
 | | Crystal structure of human glutaminase in complex with inhibitor 3 | | Descriptor: | 5,5'-pentane-1,5-diylbis(1,3,4-thiadiazol-2-amine), Glutaminase kidney isoform, mitochondrial, ... | | Authors: | Thangavelu, K, Sivaraman, J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3VP1
 
 | |
3VU6
 
 | | Short peptide HIV entry inhibitor MT-SC22EK with a M-T hook | | Descriptor: | MTSC22, SULFATE ION, Transmembrane protein gp41 | | Authors: | Yao, X, Chong, H.H, Waltersperger, S, Wang, M.T, He, Y.X, Cui, S. | | Deposit date: | 2012-06-19 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.324 Å) | | Cite: | Short-peptide fusion inhibitors with high potency against wild-type and enfuvirtide-resistant HIV-1
Faseb J., 27, 2013
|
|
3VP4
 
 | | Crystal structure of human glutaminase in complex with inhibitor 4 | | Descriptor: | 5,5'-butane-1,4-diylbis(1,3,4-thiadiazol-2-amine), Glutaminase kidney isoform, mitochondrial | | Authors: | Thangavelu, K, Sivaraman, J. | | Deposit date: | 2012-02-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZNV
 
 | | Crystal structure of the OTU domain of OTULIN at 1.3 Angstroms. | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Keusekotten, K, Elliott, P.R, Glockner, L, Kulathu, Y, Wauer, T, Krappmann, D, Hofmann, K, Komander, D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Otulin Antagonizes Lubac Signaling by Specifically Hydrolyzing met1-Linked Polyubiquitin.
Cell(Cambridge,Mass.), 153, 2013
|
|
3VU5
 
 | | Short peptide HIV entry Inhibitor SC22EK | | Descriptor: | AMMONIUM ION, SC22, SULFATE ION, ... | | Authors: | Yao, X, Chong, H.H, Waltersperger, S, Wang, M.T, He, Y.X, Cui, S. | | Deposit date: | 2012-06-19 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.087 Å) | | Cite: | Short-peptide fusion inhibitors with high potency against wild-type and enfuvirtide-resistant HIV-1
Faseb J., 27, 2013
|
|
4JYT
 
 | | Crystal Structure of Matriptase in complex with Inhibitor | | Descriptor: | 4,4'-[{3-[(naphthalen-2-ylsulfonyl)amino]pyridine-2,6-diyl}bis(oxy)]dibenzenecarboximidamide, Suppressor of tumorigenicity 14 protein | | Authors: | Subramanya, H.S, Ravi, B.C, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S. | | Deposit date: | 2013-04-01 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Pyridyl Bis(oxy)dibenzimidamide Derivatives as Selective Matriptase Inhibitors
ACS MED.CHEM.LETT., 4, 2013
|
|
4JZ1
 
 | | Crystal Structure of Matriptase in complex with Inhibitor | | Descriptor: | 4,4'-[(3-{[(4-fluorophenyl)sulfonyl]amino}pyridine-2,6-diyl)bis(oxy)]dibenzenecarboximidamide, Suppressor of tumorigenicity 14 protein | | Authors: | Subramanya, H.S, Ravi, B.C, Ashok, K.N, Chakshusmathi, G, Ramesh, K.S. | | Deposit date: | 2013-04-02 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Pyridyl Bis(oxy)dibenzimidamide Derivatives as Selective Matriptase Inhibitors
ACS MED.CHEM.LETT., 4, 2013
|
|
