7QKB
 
 | | Crystal structure of human Cathepsin L in complex with covalently bound GC376 | | Descriptor: | CHLORIDE ION, Cathepsin L, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Reinke, P.Y.A, Falke, S, Lieske, J, Ewert, W, Loboda, J, Rahmani Mashhour, A, Hauser, M, Karnicar, K, Usenik, A, Lindic, N, Lach, M, Boehler, H, Beck, T, Cox, R, Chapman, H.N, Hinrichs, W, Turk, D, Guenther, S, Meents, A. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Calpeptin is a potent cathepsin inhibitor and drug candidate for SARS-CoV-2 infections.
Commun Biol, 6, 2023
|
|
1OS8
 
 | | RECOMBINANT STREPTOMYCES GRISEUS TRYPSIN | | Descriptor: | CALCIUM ION, SULFATE ION, trypsin | | Authors: | Page, M.J, Wong, S.L, Hewitt, J, Strynadka, N.C, MacGillivray, R.T. | | Deposit date: | 2003-03-18 | | Release date: | 2003-08-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Engineering the Primary Substrate Specificity of Streptomyces griseus Trypsin.
Biochemistry, 42, 2003
|
|
6Y6K
 
 | | Cryo-EM structure of a Phenuiviridae L protein | | Descriptor: | MAGNESIUM ION, RNA-dependent RNA polymerase | | Authors: | Vogel, D, Thorkelsson, S.R, Quemin, E, Meier, K, Kouba, T, Gogrefe, N, Busch, C, Reindl, S, Guenther, S, Cusack, S, Gruenewald, K, Rosenthal, M. | | Deposit date: | 2020-02-26 | | Release date: | 2020-04-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural and functional characterization of the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 48, 2020
|
|
5ZYR
 
 | | Crystal structure of the reductase (C1) component of p-hydroxyphenylacetate 3-hydroxylase (HPAH) from Acinetobacter baumannii | | Descriptor: | ACETATE ION, FLAVIN MONONUCLEOTIDE, p-hydroxyphenylacetate 3-hydroxylase, ... | | Authors: | Oonanant, W, Phongsak, T, Sucharitakul, J, Chaiyen, P, Yuvaniyama, J. | | Deposit date: | 2018-05-28 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.20001316 Å) | | Cite: | Crystal structure of the reductase (C1) component of p-hydroxyphenylacetate 3-hydroxylase (HPAH) from Acinetobacter baumannii
To Be Published
|
|
8G4X
 
 | | Native GABA-A receptor from the mouse brain, meta-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA and allopregnanolone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8G5F
 
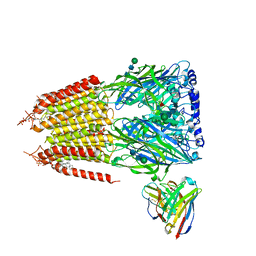 | | Native GABA-A receptor from the mouse brain, ortho-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA and allopregnanolone | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-13 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8G5H
 
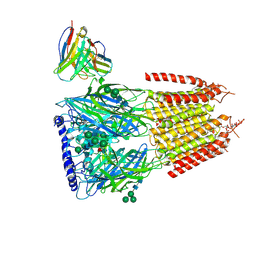 | | Native GABA-A receptor from the mouse brain, ortho-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA, Zolpidem, and endogenous neurosteroid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-13 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8G4O
 
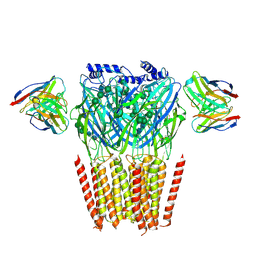 | | Native GABA-A receptor from the mouse brain, alpha1-beta2-gamma2 subtype, in complex with didesethylflurazepam and endogenous GABA | | Descriptor: | (5M)-1-(2-aminoethyl)-7-chloro-5-(2-fluorophenyl)-1,3-dihydro-2H-1,4-benzodiazepin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-10 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
8G5G
 
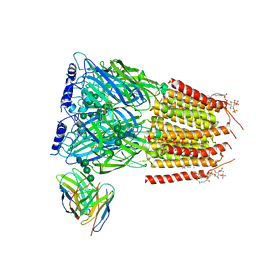 | | Native GABA-A receptor from the mouse brain, meta-alpha1-alpha3-beta2-gamma2 subtype, in complex with GABA, Zolpidem, and endogenous neurosteroid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Sun, C, Gouaux, E. | | Deposit date: | 2023-02-13 | | Release date: | 2023-09-20 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Cryo-EM structures reveal native GABA A receptor assemblies and pharmacology.
Nature, 622, 2023
|
|
5N7L
 
 | |
8EUO
 
 | | Hydroxynitrile Lyase from Hevea brasiliensis with Seven Mutations | | Descriptor: | (S)-hydroxynitrile lyase | | Authors: | Greenberg, L.R, Walsh, M.E, Kazlauskas, R.J, Pierce, C.T, Shi, K, Aihara, H, Evans, R.L. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | to be published
To Be Published
|
|
7POI
 
 | | Prodomain bound BMP10 crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bone morphogenetic protein 10, D(-)-TARTARIC ACID | | Authors: | Guo, J, Yu, M, Li, W. | | Deposit date: | 2021-09-09 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of BMPRII extracellular domain in binary and ternary receptor complexes with BMP10.
Nat Commun, 13, 2022
|
|
7POJ
 
 | | Prodomain bound BMP10 crystal form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Bone morphogenetic protein 10, TETRAETHYLENE GLYCOL | | Authors: | Guo, J, Yu, M, Li, W. | | Deposit date: | 2021-09-09 | | Release date: | 2022-05-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of BMPRII extracellular domain in binary and ternary receptor complexes with BMP10.
Nat Commun, 13, 2022
|
|
7PPB
 
 | |
6TYF
 
 | | Crystal structure of MTB sigma L transcription initiation complex with 6 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*GP*GP*T)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6TYG
 
 | | Crystal structure of MTB sigma L transcription initiation complex with 9 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*GP*GP*TP*G)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6U1V
 
 | | Crystal structure of acyl-ACP/acyl-CoA dehydrogenase from allylmalonyl-CoA and FK506 biosynthesis, TcsD | | Descriptor: | Acyl-CoA dehydrogenase domain-containing protein, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE | | Authors: | Blake-Hedges, J.M, Pereira, J.H, Barajas, J.F, Adams, P.D, Keasling, J.D. | | Deposit date: | 2019-08-16 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Mechanism of Regioselectivity in an Unusual Bacterial Acyl-CoA Dehydrogenase.
J.Am.Chem.Soc., 142, 2020
|
|
6U0O
 
 | | Crystal structure of a peptidoglycan release complex, SagB-SpdC, in lipidic cubic phase | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(2-ETHOXYETHOXY)ETHANOL, CITRATE ANION, ... | | Authors: | Owens, T.W, Schaefer, K, Kahne, D, Walker, S. | | Deposit date: | 2019-08-14 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and reconstitution of a hydrolase complex that may release peptidoglycan from the membrane after polymerization.
Nat Microbiol, 6, 2021
|
|
6TYE
 
 | | Crystal structure of MTB sigma L transcription initiation complex with 5 nt long RNA primer | | Descriptor: | DNA (5'-D(*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*AP*TP*CP*GP*AP*GP*G)-3'), DNA (5'-D(P*GP*TP*GP*TP*CP*AP*GP*TP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*C)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Molodtsov, V, Ebright, R.H. | | Deposit date: | 2019-08-08 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | RNA extension drives a stepwise displacement of an initiation-factor structural module in initial transcription.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6UP0
 
 | |
4TVX
 
 | | Crystal structure of the E. coli CRISPR RNA-guided surveillance complex, Cascade | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Jackson, R.N, Golden, S.M, Carter, J, Wiedenheft, B. | | Deposit date: | 2014-06-28 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli.
Science, 345, 2014
|
|
7C7J
 
 | |
7C7I
 
 | |
2FMJ
 
 | |
5I8F
 
 | | Crystal structure of St. John's wort Hyp-1 protein in complex with melatonin | | Descriptor: | GLYCEROL, N-[2-(5-methoxy-1H-indol-3-yl)ethyl]acetamide, Phenolic oxidative coupling protein, ... | | Authors: | Sliwiak, J, Dauter, Z, Jaskolski, M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal Structure of Hyp-1, a Hypericum perforatum PR-10 Protein, in Complex with Melatonin.
Front Plant Sci, 7, 2016
|
|
