7XV6
 
 | |
6LBR
 
 | | Crystal structure of yeast Cdc13 and ssDNA | | Descriptor: | KLLA0F20922p, Telomere single-strand DNA | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7XVA
 
 | |
7XV9
 
 | | Crystal structure of the Human TR4 DNA-Binding Domain | | Descriptor: | Nuclear receptor subfamily 2 group C member 2, ZINC ION | | Authors: | Liu, Y, Chen, Z. | | Deposit date: | 2022-05-21 | | Release date: | 2022-12-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structures of human TR4LBD-JAZF1 and TR4DBD-DNA complexes reveal the molecular basis of transcriptional regulation.
Nucleic Acids Res., 51, 2023
|
|
6LBT
 
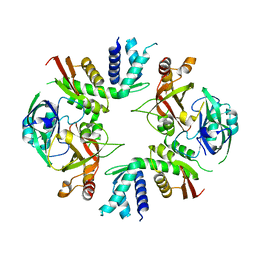 | | Crystal structure of yeast Cdc13 and Stn1 | | Descriptor: | GLYCEROL, KLLA0C11825p, KLLA0F20922p,KLLA0F20922p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6LBU
 
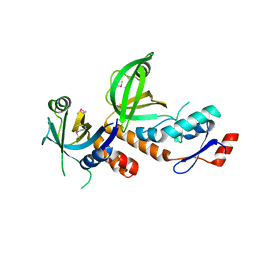 | | Crystal structure of yeast Stn1 and Ten1 | | Descriptor: | KLLA0C11825p, KLLA0E09417p | | Authors: | Ge, Y, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2019-11-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5GSA
 
 | | EED in complex with an allosteric PRC2 inhibitor | | Descriptor: | Histone-lysine N-methyltransferase EZH2, N-(furan-2-ylmethyl)-8-(4-methylsulfonylphenyl)-[1,2,4]triazolo[4,3-c]pyrimidin-5-amine, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of
Nat. Chem. Biol., 13, 2017
|
|
5H13
 
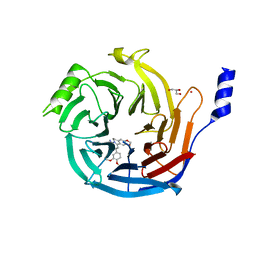 | | EED in complex with PRC2 allosteric inhibitor EED396 | | Descriptor: | 4-azanylidene-2-(3-methoxy-4-propan-2-yloxy-phenyl)-6,7-dihydro-[1,3]benzodioxolo[6,5-a]quinolizine-3-carbonitrile, GLYCEROL, PRASEODYMIUM ION, ... | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
5H17
 
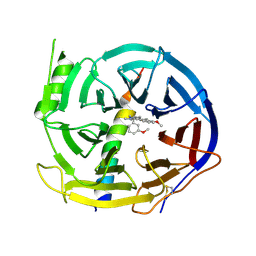 | | EED in complex with PRC2 allosteric inhibitor EED210 | | Descriptor: | (3R,4aS,10aS)-6-methoxy-3-[(3-methoxyphenyl)methyl]-1-methyl-3,4,4a,5,10,10a-hexahydro-2H-benzo[g]quinoline, Histone-lysine N-methyltransferase EZH2, Polycomb protein EED | | Authors: | Zhao, K, Zhao, M, Luo, X, Zhang, H. | | Deposit date: | 2016-10-08 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and Molecular Basis of a Diverse Set of Polycomb Repressive Complex 2 Inhibitors Recognition by EED
PLoS ONE, 12, 2017
|
|
8GUE
 
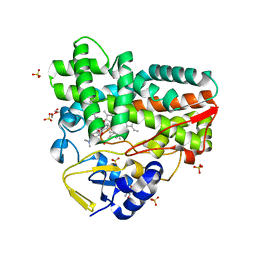 | | Crystal Structure of narbomycin-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome P450 monooxygenase PikC, ... | | Authors: | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | Deposit date: | 2022-09-11 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unnatural activities and mechanistic insights of cytochrome P450 PikC gained from site-specific mutagenesis by non-canonical amino acids.
Nat Commun, 14, 2023
|
|
7UYB
 
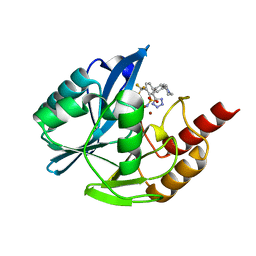 | | Inhibitor bound VIM1 | | Descriptor: | (2M)-4'-(piperidin-4-yl)-2-(1H-tetrazol-5-yl)-4-(trifluoromethyl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYC
 
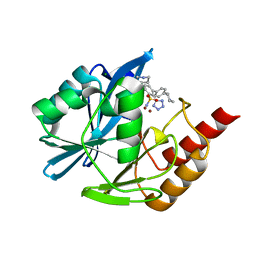 | | Inhibitor bound VIM1 | | Descriptor: | (2P)-4'-(piperidin-4-yl)-4-[(piperidin-4-yl)methyl]-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, MAGNESIUM ION, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7UYD
 
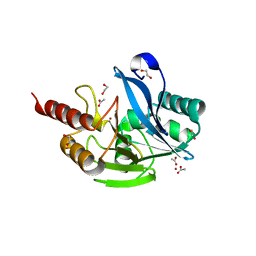 | | Inhibitor bound VIM1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Metallo-beta-lactamase VIM-2-like protein, ... | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
1X79
 
 | | Crystal structure of human GGA1 GAT domain complexed with the GAT-binding domain of Rabaptin5 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ADP-ribosylation factor binding protein GGA1, Rab GTPase binding effector protein 1, ... | | Authors: | Zhu, G, Zhang, X.C. | | Deposit date: | 2004-08-13 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal structure of human GGA1 GAT domain complexed with the GAT-binding domain of Rabaptin5.
EMBO J., 23, 2004
|
|
4YP8
 
 | | Irak4-inhibitor co-structure | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-{1-(4-cyclopropyl-2-fluorophenyl)-3-[1-(propan-2-yl)piperidin-4-yl]-1H-pyrazol-5-yl}pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-03-12 | | Release date: | 2015-05-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.641 Å) | | Cite: | Potent and Selective Amidopyrazole Inhibitors of IRAK4 That Are Efficacious in a Rodent Model of Inflammation.
Acs Med.Chem.Lett., 6, 2015
|
|
4YO6
 
 | | Irak4-inhibitor co-structure | | Descriptor: | Interleukin-1 receptor-associated kinase 4, N-(3-methyl-1-phenyl-1H-pyrazol-5-yl)pyrazolo[1,5-a]pyrimidine-3-carboxamide | | Authors: | Fischmann, T.O. | | Deposit date: | 2015-03-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Potent and Selective Amidopyrazole Inhibitors of IRAK4 That Are Efficacious in a Rodent Model of Inflammation.
Acs Med.Chem.Lett., 6, 2015
|
|
7UYA
 
 | | Inhibitor bound VIM1 | | Descriptor: | (2M)-4'-(piperidin-4-yl)-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Fischmann, T.O, Scapin, G. | | Deposit date: | 2022-05-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Structure Guided Discovery of Novel Pan Metallo-beta-Lactamase Inhibitors with Improved Gram-Negative Bacterial Cell Penetration.
J.Med.Chem., 67, 2024
|
|
7KPV
 
 | | Structure of kinase and Central lobes of yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12, Mediator of RNA polymerase II transcription subunit 13, Meiotic mRNA stability protein kinase SSN3, ... | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
6IEG
 
 | | Crystal structure of human MTR4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Exosome RNA helicase MTR4, MAGNESIUM ION | | Authors: | Chen, J.Y, Yun, C.H. | | Deposit date: | 2018-09-14 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | NRDE2 negatively regulates exosome functions by inhibiting MTR4 recruitment and exosome interaction.
Genes Dev., 33, 2019
|
|
6IEH
 
 | | Crystal structures of the hMTR4-NRDE2 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Exosome RNA helicase MTR4, ... | | Authors: | Chen, J.Y, Yun, C.H. | | Deposit date: | 2018-09-14 | | Release date: | 2019-04-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.892 Å) | | Cite: | NRDE2 negatively regulates exosome functions by inhibiting MTR4 recruitment and exosome interaction.
Genes Dev., 33, 2019
|
|
8H1T
 
 | | Cryo-EM structure of BAP1-ASXL1 bound to chromatosome | | Descriptor: | DNA (187-MER), Histone H1.4, Histone H2A type 1-D, ... | | Authors: | Ge, W, Yu, C, Xu, R.M. | | Deposit date: | 2022-10-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Basis of the H2AK119 specificity of the Polycomb repressive deubiquitinase.
Nature, 616, 2023
|
|
3V43
 
 | | Crystal structure of MOZ | | Descriptor: | ACETATE ION, Histone H3.1, Histone acetyltransferase KAT6A, ... | | Authors: | Qiu, Y, Li, F. | | Deposit date: | 2011-12-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Combinatorial readout of unmodified H3R2 and acetylated H3K14 by the tandem PHD finger of MOZ reveals a regulatory mechanism for HOXA9 transcription
Genes Dev., 26, 2012
|
|
4NXL
 
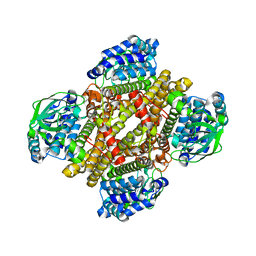 | | Dibenzothiophene monooxygenase (DszC) from Rhodococcus erythropolis | | Descriptor: | DszC | | Authors: | Zhang, L, Duan, X, Li, X, Rao, Z. | | Deposit date: | 2013-12-09 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the stabilization of active, tetrameric DszC by its C-terminus.
Proteins, 82, 2014
|
|
7KPX
 
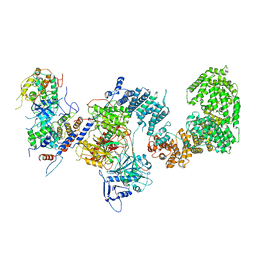 | | Structure of the yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12, Mediator of RNA polymerase II transcription subunit 13, Meiotic mRNA stability protein kinase SSN3, ... | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
7KPW
 
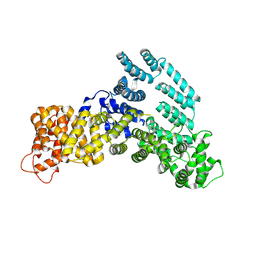 | | Structure of the H-lobe of yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12 | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
