1Q1G
 
 | | Crystal structure of Plasmodium falciparum PNP with 5'-methylthio-immucillin-H | | Descriptor: | 3,4-DIHYDROXY-2-[(METHYLSULFANYL)METHYL]-5-(4-OXO-4,5-DIHYDRO-3H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)PYRROLIDINIUM, ISOPROPYL ALCOHOL, SULFATE ION, ... | | Authors: | Shi, W, Ting, L.M, Kicska, G.A, Lewandowicz, A, Tyler, P.C, Evans, G.B, Furneaux, R.H, Kim, K, Almo, S.C, Schramm, V.L. | | Deposit date: | 2003-07-19 | | Release date: | 2004-03-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Plasmodium falciparum Purine Nucleoside Phosphorylase: CRYSTAL STRUCTURES, IMMUCILLIN INHIBITORS, AND DUAL CATALYTIC FUNCTION.
J.Biol.Chem., 279, 2004
|
|
1RSZ
 
 | | Structure of human purine nucleoside phosphorylase in complex with DADMe-Immucillin-H and sulfate | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Shi, W, Lewandowicz, A, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | Deposit date: | 2003-12-10 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural comparison of human and malarial purine nucleoside phosphorylases
To be Published
|
|
1RR6
 
 | | Structure of human purine nucleoside phosphorylase in complex with Immucillin-H and phosphate | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Shi, W, Lewandowicz, A, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | Deposit date: | 2003-12-08 | | Release date: | 2005-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Plasmodium falciparum purine nucleoside phosphorylase: crystal structures, immucillin inhibitors, and dual catalytic function.
J.Biol.Chem., 279, 2004
|
|
1RT9
 
 | | Structure of human purine nucleoside phosphorylase in complex with Immucillin-H and sulfate | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Shi, W, Lewandowicz, A, Tyler, P.C, Furneaux, R.H, Almo, S.C, Schramm, V.L. | | Deposit date: | 2003-12-10 | | Release date: | 2005-02-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural comparison of human and malarial purine nucleoside phosphorylases
To be Published
|
|
5TC5
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with butylthio-DADMe-Immucillin-A and chloride | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, CHLORIDE ION, S-methyl-5'-thioadenosine phosphorylase | | Authors: | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | TBA
To be published
|
|
5TC8
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with methylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | TBA
To be published
|
|
5TC6
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with propylthio-immucillin-A | | Descriptor: | (2S,3S,4R,5S)-2-(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)-5-[(propylsulfanyl)methyl]pyrrolidine-3,4-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | TBA
To be published
|
|
5TC7
 
 | | Crystal structure of human 5'-deoxy-5'-methylthioadenosine phosphorylase in complex with 5'-methylthiotubercidin at 1.75 angstrom | | Descriptor: | 2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-METHYLSULFANYLMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, CHLORIDE ION, PHOSPHATE ION, ... | | Authors: | Cameron, S.A, Firestone, R.S, Schramm, V.L, Almo, S.C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | TBA
To be published
|
|
3FZ0
 
 | | Inosine-Guanosine Nucleoside Hydrolase (IG-NH) | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Nucleoside hydrolase, ... | | Authors: | Vandemeulebroucke, A, Minici, C, Bruno, I, Muzzolini, L, Tornaghi, P, Parkin, D.W, Schramm, V.L, Versees, W, Steyaert, J, Degano, M. | | Deposit date: | 2009-01-23 | | Release date: | 2010-01-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and mechanism of the 6-oxopurine nucleosidase from Trypanosoma brucei brucei
Biochemistry, 49, 2010
|
|
5UGF
 
 | | Crystal structure of human purine nucleoside phosphorylase (F159Y) mutant complexed with DADMe-ImmG and phosphate | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Harijan, R.K, Cameron, S.A, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-01-08 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Catalytic-site design for inverse heavy-enzyme isotope effects in human purine nucleoside phosphorylase.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5U99
 
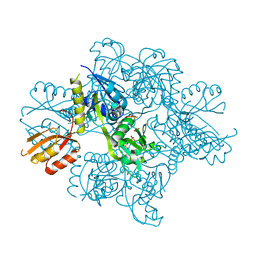 | | Transition state analysis of adenosine triphosphate phosphoribosyltransferase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP phosphoribosyltransferase, MAGNESIUM ION, ... | | Authors: | Moggre, G.-J, Poulin, M.B, Tyler, P.C, Schramm, V.L, Parker, E.J. | | Deposit date: | 2016-12-15 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Transition State Analysis of Adenosine Triphosphate Phosphoribosyltransferase.
ACS Chem. Biol., 12, 2017
|
|
7L1A
 
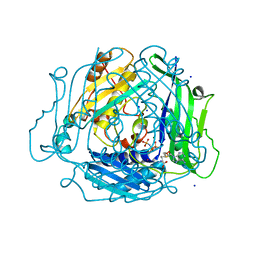 | | Human Methionine Adenosyltransferase 2A bound to Methylthioadenosine and inhibitor, di-imido triphosphate (PNPNP) | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, ALANINE, MAGNESIUM ION, ... | | Authors: | Niland, C.N, Fedorov, E, Schramm, V.L, Ghosh, A. | | Deposit date: | 2020-12-14 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Mechanism and Inhibition of Human Methionine Adenosyltransferase 2A.
Biochemistry, 60, 2021
|
|
5DK6
 
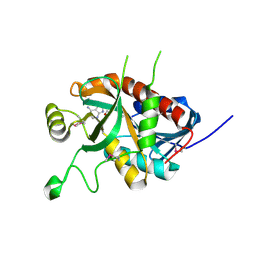 | | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH) NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION | | Descriptor: | 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ADENINE, GLYCINE | | Authors: | Himmel, D.M, Bhosle, R, Toro, R, Ahmed, M, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R.D, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-09-03 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | CRYSTAL STRUCTURE OF A 5'-METHYLTHIOADENOSINE/S-ADENOSYLHOMOCYSTEINE (MTA/SAH)NUCLEOSIDASE (MTAN) FROM COLWELLIA PSYCHRERYTHRAEA 34H (CPS_4743, TARGET PSI-029300) IN COMPLEX WITH ADENINE AT 2.27 A RESOLUTION
To be published
|
|
1Q8F
 
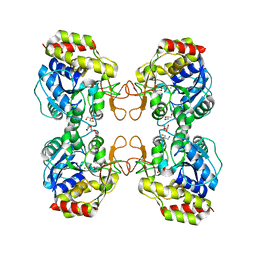 | |
8SWS
 
 | | Structure of K. lactis PNP S42E-H98R variant bound to transition state analog DADMe-IMMUCILLIN G and sulfate | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, Purine nucleoside phosphorylase, SULFATE ION | | Authors: | Fedorov, E, Ghosh, A. | | Deposit date: | 2023-05-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Phosphate Binding in PNP Alters Transition-State Analogue Affinity and Subunit Cooperativity.
Biochemistry, 62, 2023
|
|
8SWP
 
 | | Structure of K. lactis PNP bound to hypoxanthine | | Descriptor: | ACETATE ION, HYPOXANTHINE, Purine nucleoside phosphorylase | | Authors: | Fedorov, E, Ghosh, A. | | Deposit date: | 2023-05-19 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphate Binding in PNP Alters Transition-State Analogue Affinity and Subunit Cooperativity.
Biochemistry, 62, 2023
|
|
8SWR
 
 | | Structure of K. lactis PNP S42E variant bound to transition state analog DADMe-IMMUCILLIN G and sulfate | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, GUANINE, Purine nucleoside phosphorylase, ... | | Authors: | Fedorov, E, Ghosh, A. | | Deposit date: | 2023-05-19 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Phosphate Binding in PNP Alters Transition-State Analogue Affinity and Subunit Cooperativity.
Biochemistry, 62, 2023
|
|
8SWU
 
 | |
8SWQ
 
 | | Structure of K. lactis PNP bound to transition state analog DADMe-IMMUCILLIN H and sulfate | | Descriptor: | 7-[[(3R,4R)-3-(hydroxymethyl)-4-oxidanyl-pyrrolidin-1-ium-1-yl]methyl]-3,5-dihydropyrrolo[3,2-d]pyrimidin-4-one, GLYCEROL, Purine nucleoside phosphorylase, ... | | Authors: | Fedorov, E, Ghosh, A. | | Deposit date: | 2023-05-19 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Phosphate Binding in PNP Alters Transition-State Analogue Affinity and Subunit Cooperativity.
Biochemistry, 62, 2023
|
|
8SWT
 
 | |
4JWT
 
 | |
4L0M
 
 | |
4LDN
 
 | |
4KN5
 
 | |
1Z3A
 
 | | Crystal structure of tRNA adenosine deaminase TadA from Escherichia coli | | Descriptor: | ZINC ION, tRNA-specific adenosine deaminase | | Authors: | Malashkevich, V, Kim, J, Lisbin, M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-03-10 | | Release date: | 2006-02-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural and kinetic characterization of Escherichia coli TadA, the wobble-specific tRNA deaminase.
Biochemistry, 45, 2006
|
|
