3NVK
 
 | | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle | | Descriptor: | 50S ribosomal protein L7Ae, Fibrillarin-like rRNA/tRNA 2'-O-methyltransferase, NOP5/NOP56 related protein, ... | | Authors: | Xue, S, Wang, R, Li, H. | | Deposit date: | 2010-07-08 | | Release date: | 2011-07-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle.
Mol.Cell, 39, 2010
|
|
3J2X
 
 | |
3J30
 
 | |
3NVM
 
 | |
6UEG
 
 | | Pseudomonas aeruginosa LpxA Complex Structure with Ligand | | Descriptor: | 3-({2-[(2R)-2-carbamoyl-2,3-dihydro-4H-1,4-benzoxazin-4-yl]-2-oxoethyl}sulfanyl)propanoic acid, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, CALCIUM ION | | Authors: | Chen, Y, Kroeck, K, Sacco, M. | | Deposit date: | 2019-09-20 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.
Sci Rep, 9, 2019
|
|
6WTT
 
 | | Crystals Structure of the SARS-CoV-2 (COVID-19) main protease with inhibitor GC-376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Sacco, M, Ma, C, Chen, Y, Wang, J. | | Deposit date: | 2020-05-03 | | Release date: | 2020-05-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease.
Cell Res., 30, 2020
|
|
6UEC
 
 | | Pseudomonas aeruginosa LpxD Complex Structure with Ligand | | Descriptor: | 4-(naphthalen-1-yl)-4-oxobutanoic acid, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Chen, Y, Kroeck, K, Sacco, M. | | Deposit date: | 2019-09-20 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.
Sci Rep, 9, 2019
|
|
6UEE
 
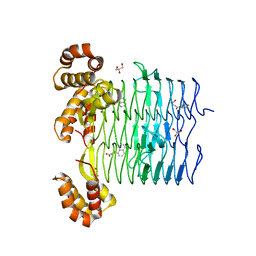 | | Pseudomonas aeruginosa LpxA Complex Structure with Ligand | | Descriptor: | 4-(naphthalen-1-yl)-4-oxobutanoic acid, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, GLYCEROL | | Authors: | Chen, Y, Kroeck, K, Sacco, M. | | Deposit date: | 2019-09-20 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.
Sci Rep, 9, 2019
|
|
6UN3
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with ticarcillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2019-10-10 | | Release date: | 2019-10-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
6UN1
 
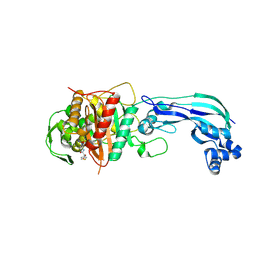 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with temocillin | | Descriptor: | (2R,4S)-2-[(1S)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-1-methoxy-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4 -carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2019-10-10 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
6VEN
 
 | |
6UUS
 
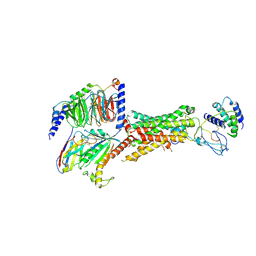 | | CryoEM Structure of the active Adrenomedullin 2 receptor G protein complex with adrenomedullin peptide | | Descriptor: | ADM, Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Liang, Y.L, Sexton, P, Danev, R. | | Deposit date: | 2019-10-31 | | Release date: | 2020-04-01 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure and Dynamics of Adrenomedullin Receptors AM1and AM2Reveal Key Mechanisms in the Control of Receptor Phenotype by Receptor Activity-Modifying Proteins.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6BU3
 
 | |
6UUN
 
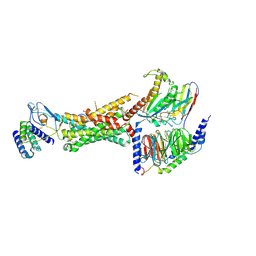 | | CryoEM Structure of the active Adrenomedullin 1 receptor G protein complex with adrenomedullin peptide | | Descriptor: | ADM, Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Liang, Y.L, Sexton, P, Danev, R. | | Deposit date: | 2019-10-30 | | Release date: | 2020-03-25 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and Dynamics of Adrenomedullin Receptors AM1and AM2Reveal Key Mechanisms in the Control of Receptor Phenotype by Receptor Activity-Modifying Proteins.
Acs Pharmacol Transl Sci, 3, 2020
|
|
6VC8
 
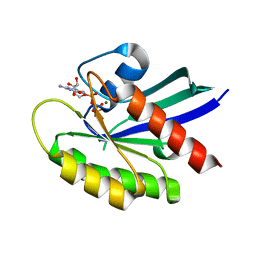 | | Crystal structure of wild-type KRAS4b(1-169) in complex with GMPPNP and Mg ion | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Tran, T.H, Davies, D.R, Edwards, T.E, Simanshu, D.K. | | Deposit date: | 2019-12-20 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Machine learning-driven multiscale modeling reveals lipid-dependent dynamics of RAS signaling proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6BT6
 
 | |
6UNB
 
 | | Crystal structure of CTX-M-14 in complex with temocillin | | Descriptor: | (2R,4S)-2-[(1S)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-1-methoxy-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4 -carboxylic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2019-10-11 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
6UED
 
 | | Apo Pseudomonas aeruginosa LpxD Structure | | Descriptor: | GLYCEROL, MAGNESIUM ION, UDP-3-O-acylglucosamine N-acyltransferase | | Authors: | Chen, Y, Kroeck, K, Sacco, M. | | Deposit date: | 2019-09-20 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of dual-activity small-molecule ligands of Pseudomonas aeruginosa LpxA and LpxD using SPR and X-ray crystallography.
Sci Rep, 9, 2019
|
|
6UVA
 
 | | CryoEM Structure of the active Adrenomedullin 2 receptor G protein complex with adrenomedullin 2 peptide | | Descriptor: | Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Belousoff, M.J, Liang, Y.L, Sexton, P, Danev, R. | | Deposit date: | 2019-11-01 | | Release date: | 2020-04-01 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structure and Dynamics of Adrenomedullin Receptors AM1and AM2Reveal Key Mechanisms in the Control of Receptor Phenotype by Receptor Activity-Modifying Proteins.
Acs Pharmacol Transl Sci, 3, 2020
|
|
2MNZ
 
 | | NMR Structure of KDM5B PHD1 finger in complex with H3K4me0(1-10aa) | | Descriptor: | H3K4me0, Lysine-specific demethylase 5B, ZINC ION | | Authors: | Zhang, Y, Yang, H.R, Guo, X, Rong, N.Y, Song, Y.J, Xu, Y.W, Lan, W.X, Xu, Y.H, Cao, C. | | Deposit date: | 2014-04-16 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B.
Protein Cell, 5, 2014
|
|
2MNY
 
 | | NMR Structure of KDM5B PHD1 finger | | Descriptor: | Lysine-specific demethylase 5B, ZINC ION | | Authors: | Zhang, Y, Yang, H.R, Guo, X, Rong, N.Y, Song, Y.J, Xu, Y.W, Lan, W.X, Xu, Y.H, Cao, C. | | Deposit date: | 2014-04-16 | | Release date: | 2014-08-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The PHD1 finger of KDM5B recognizes unmodified H3K4 during the demethylation of histone H3K4me2/3 by KDM5B.
Protein Cell, 5, 2014
|
|
1SQT
 
 | | Substituted 2-Naphthamidine Inhibitors of Urokinase | | Descriptor: | 7-METHOXY-8-[1-(METHYLSULFONYL)-1H-PYRAZOL-4-YL]NAPHTHALENE-2-CARBOXIMIDAMIDE, Urokinase-type plasminogen activator | | Authors: | Wendt, M.D, Geyer, A, McClellan, W.J, Rockway, T.W, Weitzberg, M, Zhang, X, Mantei, R, Stewart, K, Nienaber, V, Klinghofer, V, Giranda, V.L. | | Deposit date: | 2004-03-19 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interaction with the S1beta-pocket of urokinase: 8-heterocycle substituted and 6,8-disubstituted 2-naphthamidine urokinase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
2LI6
 
 | |
5YF4
 
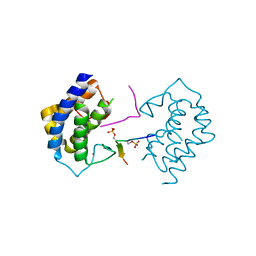 | | A kinase complex MST4-MOB4 | | Descriptor: | MOB-like protein phocein, Peptide from Serine/threonine-protein kinase 26, ZINC ION | | Authors: | Chen, M, Zhou, Z.C. | | Deposit date: | 2017-09-20 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | The MST4-MOB4 complex disrupts the MST1-MOB1 complex in the Hippo-YAP pathway and plays a pro-oncogenic role in pancreatic cancer.
J. Biol. Chem., 293, 2018
|
|
5YSW
 
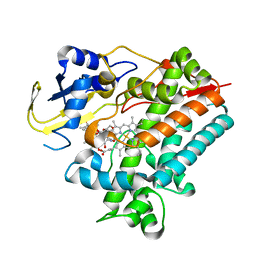 | | Crystal Structure Analysis of Rif16 in complex with R-L | | Descriptor: | (2S,12E,14E,16S,17S,18R,19R,20R,21S,22R,23S,24E)-21-(acetyloxy)-5,6,17,19-tetrahydroxy-23-methoxy-2,4,12,16,18,20,22-heptamethyl-1,11-dioxo-1,2-dihydro-2,7-(epoxypentadeca[1,11,13]trienoimino)naphtho[2,1-b]furan-9-yl hydroxyacetate, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, F.W, Qi, F.F, Xiao, Y.L, Zhao, G.P, Li, S.Y. | | Deposit date: | 2017-11-15 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Deciphering the late steps of rifamycin biosynthesis.
Nat Commun, 9, 2018
|
|
