4U63
 
 | | Crystal structure of a bacterial class III photolyase from Agrobacterium tumefaciens at 1.67A resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,10-METHENYL-6,7,8-TRIHYDROFOLIC ACID, DNA photolyase, ... | | Authors: | Scheerer, P, Zhang, F, Kalms, J, von Stetten, D, Krauss, N, Oberpichler, I, Lamparter, T. | | Deposit date: | 2014-07-26 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Class III Cyclobutane Pyrimidine Dimer Photolyase Structure Reveals a New Antenna Chromophore Binding Site and Alternative Photoreduction Pathways.
J.Biol.Chem., 290, 2015
|
|
4U64
 
 | | Structure of the periplasmic output domain of the Legionella pneumophila LapD ortholog CdgS9 in the apo state | | Descriptor: | Two component histidine kinase, GGDEF domain protein/EAL domain protein | | Authors: | Chatterjee, D, Cooley, R.B, Boyd, C.D, Mehl, R.A, O'Toole, G.A, Sondermann, H.S. | | Deposit date: | 2014-07-27 | | Release date: | 2014-08-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Mechanistic insight into the conserved allosteric regulation of periplasmic proteolysis by the signaling molecule cyclic-di-GMP.
Elife, 3, 2014
|
|
6TKG
 
 | | Tsetse thrombin inhibitor in complex with human alpha-thrombin - orthorhombic form at 12keV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Prothrombin, ... | | Authors: | Calisto, B.M, Ripoll-Rozada, J, de Sanctis, D, Pereira, P.J.B. | | Deposit date: | 2019-11-28 | | Release date: | 2020-11-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Sulfotyrosine-Mediated Recognition of Human Thrombin by a Tsetse Fly Anticoagulant Mimics Physiological Substrates.
Cell Chem Biol, 28, 2021
|
|
6TC2
 
 | | Monoclinic human insulin in complex with p-coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Insulin, PHOSPHATE ION, ... | | Authors: | Triandafillidis, D.-P, Parthenios, N, Spiliopoulou, M, Valmas, A, Kosinas, C, Gozzo, F, Reinle-Schmitt, M, Beckers, D, Degen, T, Pop, M, Fitch, A, Wollenhaupt, J, Weiss, M.S, Karavassili, F, Margiolaki, I. | | Deposit date: | 2019-11-04 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Insulin polymorphism induced by two polyphenols: new crystal forms and advances in macromolecular powder diffraction.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6T39
 
 | | Crystal structure of rsEGFP2 in its off-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Coquelle, N, Adam, V, Barends, T.R.M, De La Mora, E, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2019-10-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Photoswitching mechanism of a fluorescent protein revealed by time-resolved crystallography and transient absorption spectroscopy.
Nat Commun, 11, 2020
|
|
6TKJ
 
 | | Tsetse thrombin inhibitor in complex with human alpha-thrombin - tetragonal form at 7keV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombin heavy chain, Thrombin light chain, ... | | Authors: | Calisto, B.M, Ripoll-Rozada, J, de Sanctis, D, Pereira, P.J.B. | | Deposit date: | 2019-11-28 | | Release date: | 2020-11-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Sulfotyrosine-Mediated Recognition of Human Thrombin by a Tsetse Fly Anticoagulant Mimics Physiological Substrates.
Cell Chem Biol, 28, 2021
|
|
6TSN
 
 | | Marasmius oreades agglutinin (MOA), papain back.swap W208Q-Q276W variant | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin, CALCIUM ION, ... | | Authors: | Cordara, G, Manna, D, Krengel, U. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of MOA in complex with a peptide fragment: A protease caught in flagranti .
Curr Res Struct Biol, 2, 2020
|
|
6TSU
 
 | | Capsid of empty GTA particle computed with C5 symmetry | | Descriptor: | Major capsid protein Rcc01687, Uncharacterized protein | | Authors: | Bardy, P, Fuzik, T, Hrebik, D, Pantucek, R, Beatty, J.T, Plevka, P. | | Deposit date: | 2019-12-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structure and mechanism of DNA delivery of a gene transfer agent.
Nat Commun, 11, 2020
|
|
6TUL
 
 | | Structure of the arginase-2-inhibitory human antigen-binding fragment Fab C0021177 | | Descriptor: | D-MALATE, Fab C0021177 heavy chain (IgG1), Fab C0021177 light chain (IgG1), ... | | Authors: | Burschowsky, D, Addyman, A, Fiedler, S, Groves, M, Haynes, S, Seewooruthun, C, Carr, M. | | Deposit date: | 2020-01-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional characterization of C0021158, a high-affinity monoclonal antibody that inhibits Arginase 2 function via a novel non-competitive mechanism of action.
Mabs, 12
|
|
6TI7
 
 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
6TAY
 
 | | Mouse RNF213 mutant R4753K modeling the Moyamoya-disease-related Human variant R4810K | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNF213,E3 ubiquitin-protein ligase RNF213,E3 ubiquitin-protein ligase RNF213, ... | | Authors: | Ahel, J, Meinhart, A, Haselbach, D, Clausen, T. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Moyamoya disease factor RNF213 is a giant E3 ligase with a dynein-like core and a distinct ubiquitin-transfer mechanism.
Elife, 9, 2020
|
|
4UE4
 
 | | Structural basis for targeting and elongation arrest of Bacillus signal recognition particle | | Descriptor: | 6S RNA, FTSQ SIGNAL SEQUENCE, SIGNAL RECOGNITION PARTICLE PROTEIN | | Authors: | Beckert, B, Kedrov, A, Sohmen, D, Kempf, G, Wild, K, Sinning, I, Stahlberg, H, Wilson, D.N, Beckmann, R. | | Deposit date: | 2014-12-15 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions.
Nat.Struct.Mol.Biol., 22, 2015
|
|
6T9C
 
 | |
6TAC
 
 | | Human NAMPT deletion mutant in complex with nicotinamide mononucleotide, pyrophosphate, and Mg2+ | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Houry, D, Raasakka, A, Kursula, P, Ziegler, M. | | Deposit date: | 2019-10-29 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of structural determinants of NAMPT activity and substrate selectivity
To Be Published
|
|
3DHC
 
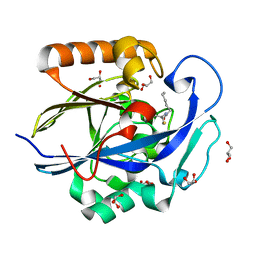 | | 1.3 Angstrom Structure of N-Acyl Homoserine Lactone Hydrolase with the Product N-Hexanoyl-L-Homocysteine Bound to The catalytic Metal Center | | Descriptor: | GLYCEROL, N-Acyl Homoserine Lactone Hydrolase, N-hexanoyl-L-homocysteine, ... | | Authors: | Liu, D, Momb, J, Thomas, P.W, Moulin, A, Petsko, G.A, Fast, W, Ringe, D. | | Deposit date: | 2008-06-17 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Mechanism of the quorum-quenching lactonase (AiiA) from Bacillus thuringiensis. 1. Product-bound structures.
Biochemistry, 47, 2008
|
|
6T3A
 
 | | Difference-refined structure of rsEGFP2 10 ns following 400-nm laser irradiation of the off-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Coquelle, N, Adam, V, Barends, T.R.M, De La Mora, E, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2019-10-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Photoswitching mechanism of a fluorescent protein revealed by time-resolved crystallography and transient absorption spectroscopy.
Nat Commun, 11, 2020
|
|
4U65
 
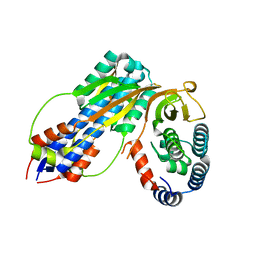 | | Structure of the periplasmic output domain of the Legionella pneumophila LapD ortholog CdgS9 in complex with Pseudomonas fluorescens LapG | | Descriptor: | CALCIUM ION, Putative cystine protease, Two component histidine kinase, ... | | Authors: | Chatterjee, D, Cooley, R.B, Boyd, C.D, Mehl, R.A, O'Toole, G.A, Sondermann, H.S. | | Deposit date: | 2014-07-27 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic insight into the conserved allosteric regulation of periplasmic proteolysis by the signaling molecule cyclic-di-GMP.
Elife, 3, 2014
|
|
6TFD
 
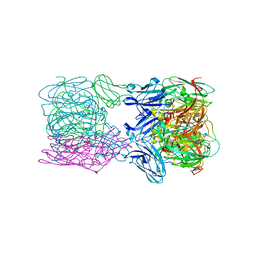 | | Crystal structure of nitrite and NO bound three-domain copper-containing nitrite reductase from Hyphomicrobium denitrificans strain 1NES1 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRIC OXIDE, ... | | Authors: | Sasaki, D, Watanabe, T.F, Eady, R.R, Garratt, R.C, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-11-13 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structures of substrate- and product-bound forms of a multi-domain copper nitrite reductase shed light on the role of domain tethering in protein complexes.
Iucrj, 7, 2020
|
|
5Q04
 
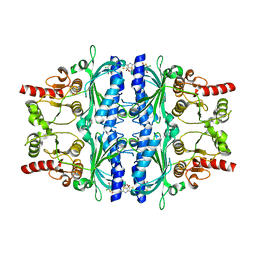 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(4-bromo-5-chlorothiophen-2-yl)sulfonyl-3-(5-bromo-1,3-thiazol-2-yl)urea | | Descriptor: | 4-bromo-N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-5-chlorothiophene-2-sulfonamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(4-bromo-5-chlorothiophen-2-yl)sulfonyl-3-(5-bromo-1,3-thiazol-2-yl)urea
To be published
|
|
6TL9
 
 | | CRYSTAL STRUCTURE OF LECTIN-LIKE OX-LDL RECEPTOR 1 IN COMPLEX WITH BI-0115 | | Descriptor: | 9-chloranyl-5-propyl-11~{H}-pyrido[2,3-b][1,4]benzodiazepin-6-one, GLYCEROL, Oxidized low-density lipoprotein receptor 1 | | Authors: | Nar, H, Fiegen, D, Schnapp, G. | | Deposit date: | 2019-12-02 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | A small-molecule inhibitor of lectin-like oxidized LDL receptor-1 acts by stabilizing an inactive receptor tetramer state
Commun Chem, 2020
|
|
4UQH
 
 | | Crystal structure of Trypanosoma cruzi CYP51 bound to the inhibitor (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(3,4-difluorophenyl)piperazin-1-yl)-2-fluorobenzamide. | | Descriptor: | (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(3,4-difluorophenyl)piperazin-1-yl)-2-fluorobenzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE, ... | | Authors: | Calvet, C.M, Vieira, D.F, Choi, J.Y, Cameron, M.D, Gut, J, Kellar, D, Siqueira-Neto, J.L, McKerrow, J.H, Roush, W.R, Podust, L.M. | | Deposit date: | 2014-06-23 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | 4-Aminopyridyl-Based Cyp51 Inhibitors as Anti-Trypanosoma Cruzi Drug Leads with Improved Pharmacokinetic Profile and in Vivo Potency.
J.Med.Chem., 57, 2014
|
|
4UMM
 
 | | The Cryo-EM structure of the palindromic DNA-bound USP-EcR nuclear receptor reveals an asymmetric organization with allosteric domain positioning | | Descriptor: | 2,3,14,20,22-PENTAHYDROXYCHOLEST-7-EN-6-ONE, 5'-D(*CP*AP*AP*GP*GP*GP*TP*TP*CP*AP*AP*TP*GP*CP *AP*CP*TP*TP*GP*TP)-3', 5'-D(*DGP*AP*CP*AP*AP*GP*TP*GP*CP*AP*TP*TP*GP*DAP *AP*CP*CP*CP*TP*T)-3', ... | | Authors: | Maletta, M, Orlov, I, Moras, D, Billas, I.M.L, Klaholz, B.P. | | Deposit date: | 2014-05-19 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11.6 Å) | | Cite: | The Palindromic DNA-Bound Usp-Ecr Nuclear Receptor Adopts an Asymmetric Organization with Allosteric Domain Positioning.
Nat.Commun., 5, 2014
|
|
4UJ6
 
 | | Structure of surface layer protein SbsC, domains 1-6 | | Descriptor: | SURFACE LAYER PROTEIN | | Authors: | Dordic, A, Pavkov-Keller, T, Eder, M, Egelseer, E.M, Davis, K, Mills, D, Sleytr, U.B, Kuehlbrandt, W, Vonck, J, Keller, W. | | Deposit date: | 2015-04-08 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure of Surface Layer Protein Sbsc, Domains 1-6
To be Published
|
|
6TSP
 
 | | Marasmius oreades agglutinin (MOA) inhibited by zinc | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin, CALCIUM ION, ... | | Authors: | Cordara, G, Manna, D, Krengel, U. | | Deposit date: | 2019-12-21 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of MOA in complex with a peptide fragment: a protease caught in flagranti
Curr Res Struct Biol, 2, 2020
|
|
6T5J
 
 | | Structure of NUDT15 in complex with inhibitor TH1760 | | Descriptor: | 6-[4-(1~{H}-indol-5-ylcarbonyl)piperazin-1-yl]sulfonyl-3~{H}-1,3-benzoxazol-2-one, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Carter, M, Rehling, D, Desroses, M, Zhang, S.M, Hagenkort, A, Valerie, N.C.K, Helleday, T, Stenmark, P. | | Deposit date: | 2019-10-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of a chemical probe against NUDT15.
Nat.Chem.Biol., 16, 2020
|
|
