7WKU
 
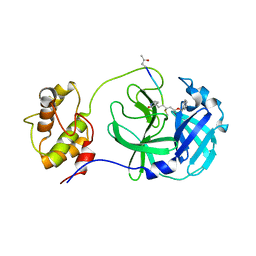 | | Structure of PDCoV Mpro in complex with an inhibitor | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, Peptidase C30 | | Authors: | Wang, F.H, Yang, H.T. | | Deposit date: | 2022-01-11 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of the Porcine Deltacoronavirus Main Protease Reveals a Conserved Target for the Design of Antivirals.
Viruses, 14, 2022
|
|
7Y3Y
 
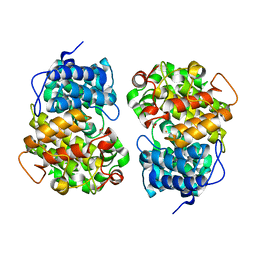 | | Crystal structure of BTG13 mutant (T299V) | | Descriptor: | FE (III) ION, GLYCEROL, questin oxidase BTG13 | | Authors: | Hou, X.D, Fu, K, Rao, Y.J. | | Deposit date: | 2022-06-13 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Discovery of the Biosynthetic Pathway of Beticolin 1 Reveals a Novel Non-Heme Iron-Dependent Oxygenase for Anthraquinone Ring Cleavage.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
7Y3X
 
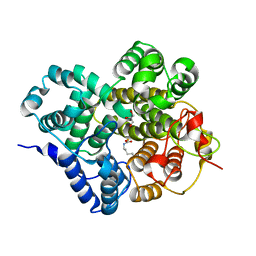 | | Crystal structure of BTG13 mutant (H58F) | | Descriptor: | FE (III) ION, GLYCEROL, Questin oxidase | | Authors: | Hou, X.D, Rao, Y.J. | | Deposit date: | 2022-06-13 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery of the Biosynthetic Pathway of Beticolin 1 Reveals a Novel Non-Heme Iron-Dependent Oxygenase for Anthraquinone Ring Cleavage.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
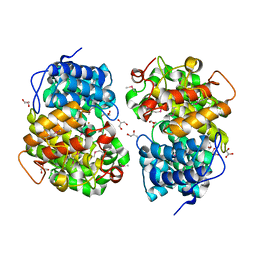
7Y3W
 
 | |
8JMT
 
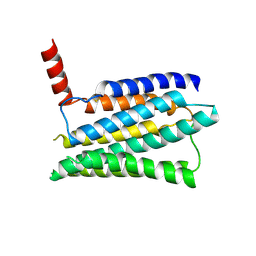 | | Structure of the adhesion GPCR ADGRL3 in the apo state | | Descriptor: | Adhesion G protein-coupled receptor L3,Soluble cytochrome b562 | | Authors: | Tao, Y, Guo, Q, He, B, Zhong, Y. | | Deposit date: | 2023-06-05 | | Release date: | 2023-09-06 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8JJ8
 
 | | Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with a partial agonist | | Descriptor: | Beta-2 adrenergic receptor,Soluble cytochrome b562, ~{N}-[5-[(1~{R})-2-[[(2~{R})-1-(4-methoxyphenyl)propan-2-yl]amino]-1-oxidanyl-ethyl]-2-oxidanyl-phenyl]methanamide | | Authors: | He, B.B, Zhong, Y.X, Guo, Q, Tao, Y.Y. | | Deposit date: | 2023-05-29 | | Release date: | 2023-09-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8JJL
 
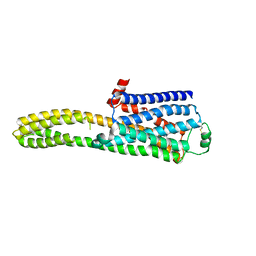 | | cryo-EM structure of the beta2-AR-mBRIL/1b3 Fab/Glue complex with a full agonist | | Descriptor: | Beta-2 adrenergic receptor,Soluble cytochrome b562, Olodaterol | | Authors: | He, B.B, Zhong, Y.X, Guo, Q, Tao, Y.Y. | | Deposit date: | 2023-05-30 | | Release date: | 2023-09-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8J7E
 
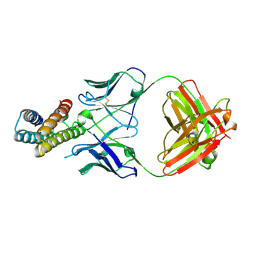 | | Crystal structure of BRIL in complex with 1b3 Fab | | Descriptor: | Antibody 1b3 Fab Heavy chain, Antibody 1b3 Fab Light chain, Soluble cytochrome b562 | | Authors: | Zhong, Y.X, Guo, Q, Tao, Y.Y. | | Deposit date: | 2023-04-27 | | Release date: | 2023-09-06 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8JJO
 
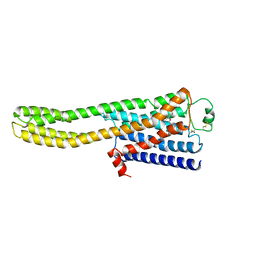 | | Cryo-EM structure of the beta2AR-mBRIL/1b3 Fab/Glue complex with an antagonist | | Descriptor: | (2S)-1-[(1-methylethyl)amino]-3-(2-prop-2-en-1-ylphenoxy)propan-2-ol, Beta-2 adrenergic receptor,Beta-2 adrenergic receptor,Soluble cytochrome b562 | | Authors: | He, B.B, Zhong, Y.X, Guo, Q, Tao, Y.Y. | | Deposit date: | 2023-05-31 | | Release date: | 2023-09-06 | | Last modified: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A method for structure determination of GPCRs in various states.
Nat.Chem.Biol., 20, 2024
|
|
8KIF
 
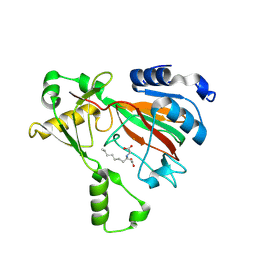 | | The structure of MmaE with substrate | | Descriptor: | (3R)-3-(2-hydroxy-2-oxoethylamino)decanoic acid, FE (II) ION, Putative dioxygenase | | Authors: | Chen, J, Zhou, J. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Variation in biosynthesis and metal-binding properties of isonitrile-containing peptides produced by Mycobacteria versus Streptomyces.
Acs Catalysis, 14, 2024
|
|
8KHT
 
 | | The structure of Rv0097 with substrate | | Descriptor: | (3R)-3-(2-hydroxy-2-oxoethylamino)decanoic acid, FE (II) ION, Oxidoreductase | | Authors: | Chen, J, Zhou, J. | | Deposit date: | 2023-08-22 | | Release date: | 2024-04-17 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Variation in biosynthesis and metal-binding properties of isonitrile-containing peptides produced by Mycobacteria versus Streptomyces.
Acs Catalysis, 14, 2024
|
|
7X4B
 
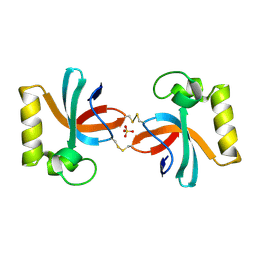 | | Crystal Structure of An Anti-CRISPR Protein | | Descriptor: | Anti-CRISPR protein (AcrIIC1), SULFATE ION | | Authors: | Hu, J, Zhang, S, Gao, J.Y, Liu, X, Liu, J. | | Deposit date: | 2022-03-02 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | A redox switch regulates the assembly and anti-CRISPR activity of AcrIIC1.
Nat Commun, 13, 2022
|
|
7VW5
 
 | |
7VB4
 
 | |
7XJP
 
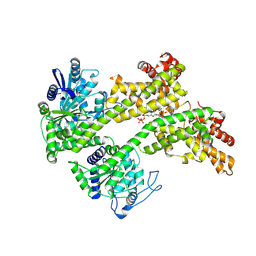 | | Cryo-EM structure of EDS1 and SAG101 with ATP-APDR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ADENOSINE-5-DIPHOSPHORIBOSE, ISOPROPYL ALCOHOL, ... | | Authors: | Huang, S.J, Jia, A.L, Han, Z.F, Chai, J.J. | | Deposit date: | 2022-04-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity.
Science, 377, 2022
|
|
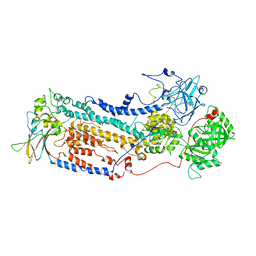
7VGI
 
 | |
7VGJ
 
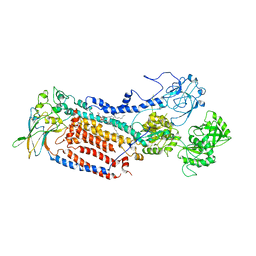 | | Cryo-EM structure of the human P4-type flippase ATP8B1-CDC50A in the auto-inhibited E2Pi-PS state | | Descriptor: | Cell cycle control protein 50A, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, Phospholipid-transporting ATPase IC | | Authors: | Chen, M.T, Chen, Y, Chen, Z.P, Zhou, C.Z, Hou, W.T, Chen, Y. | | Deposit date: | 2021-09-16 | | Release date: | 2022-03-30 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structural insights into the activation of autoinhibited human lipid flippase ATP8B1 upon substrate binding.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
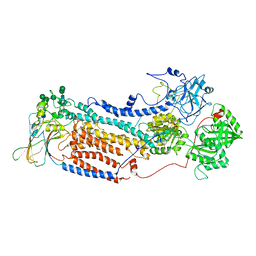
7VGH
 
 | |
7XME
 
 | | Structure of Influenza A virus polymerase basic protein 2 (PB2) with an azazindole derivative | | Descriptor: | (2~{S},3~{S})-3-[[5-dimethoxyphosphoryl-4-(5-fluoranyl-1~{H}-pyrrolo[2,3-b]pyridin-3-yl)pyrimidin-2-yl]amino]bicyclo[2.2.2]octane-2-carboxylic acid, IODIDE ION, Polymerase basic protein 2 | | Authors: | Zhang, Z. | | Deposit date: | 2022-04-25 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.521 Å) | | Cite: | Discovery of a novel azaindole derivatives targeting the influenza PB2 cap binding region
To Be Published
|
|
7XOZ
 
 | | Crystal structure of RPPT-TIR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase | | Authors: | Song, W, Jia, A, Huang, S, Chai, J. | | Deposit date: | 2022-05-02 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | TIR-catalyzed ADP-ribosylation reactions produce signaling molecules for plant immunity.
Science, 377, 2022
|
|
7W3R
 
 | | USP34 catalytic domain | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 34, ZINC ION | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural Insights into the Catalytic Mechanism and Ubiquitin Recognition of USP34.
J.Mol.Biol., 434, 2022
|
|
7W3U
 
 | | USP34 catalytic domain in complex with UbPA | | Descriptor: | Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 34, ZINC ION, ... | | Authors: | Xu, G.L, Ming, Z.H. | | Deposit date: | 2021-11-26 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Structural Insights into the Catalytic Mechanism and Ubiquitin Recognition of USP34.
J.Mol.Biol., 434, 2022
|
|
7UED
 
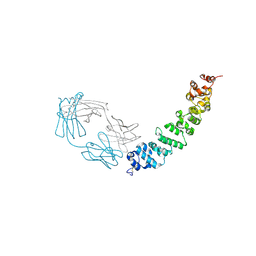 | | Crystal structure of full length mesothelin bound with MORAb-009 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Isoform 4 of Mesothelin, ... | | Authors: | Zhan, J, Esser, L, Lin, D, Tang, W.K, Xia, D. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of Cancer Antigen Mesothelin and Its Complexes with Therapeutic Antibodies.
Cancer Res Commun, 3, 2023
|
|
7D1X
 
 | |
