1FUO
 
 | | FUMARASE C WITH BOUND CITRATE | | Descriptor: | CITRIC ACID, D-MALATE, FUMARASE C | | Authors: | Weaver, T, Banaszak, L. | | Deposit date: | 1996-08-29 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystallographic studies of the catalytic and a second site in fumarase C from Escherichia coli.
Biochemistry, 35, 1996
|
|
3V0H
 
 | | Crystal structure of Ciona intestinalis voltage sensor-containing phosphatase (Ci-VSP), residues 241-576(C363S), complexed with D-MYO-inositol-1,4,5-triphosphate | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Voltage-sensor containing phosphatase | | Authors: | Liu, L, Kohout, S.C, Xu, Q, Muller, S, Kimberlin, C, Isacoff, E.Y, Minor, D.L. | | Deposit date: | 2011-12-08 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A glutamate switch controls voltage-sensitive phosphatase function.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6G1H
 
 | | Amine Dehydrogenase from Petrotoga mobilis; open form | | Descriptor: | 1,2-ETHANEDIOL, Dihydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Beloti, L, Frese, A, Mayol, O, Vergne-Vaxelaire, C, Grogan, G. | | Deposit date: | 2018-03-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | A family of native amine dehydrogenases for the asymmetric reductive amination of ketones
Nat Catal, 2019
|
|
8S8J
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-eIF5) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
1PJ3
 
 | | Crystal structure of human mitochondrial NAD(P)+-dependent malic enzyme in a pentary complex with natural substrate pyruvate, cofactor NAD+, Mn++, and allosteric activator fumarate. | | Descriptor: | FUMARIC ACID, MANGANESE (II) ION, NAD-dependent malic enzyme, ... | | Authors: | Tao, X, Yang, Z, Tong, L. | | Deposit date: | 2003-05-30 | | Release date: | 2003-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism.
Structure, 11, 2003
|
|
1FZQ
 
 | | CRYSTAL STRUCTURE OF MURINE ARL3-GDP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ADP-RIBOSYLATION FACTOR-LIKE PROTEIN 3, AMMONIUM ION, ... | | Authors: | Hillig, R.C, Hanzal-Bayer, M, Linari, M, Becker, J, Wittinghofer, A, Renault, L. | | Deposit date: | 2000-10-04 | | Release date: | 2000-12-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical properties show ARL3-GDP as a distinct GTP binding protein.
Structure Fold.Des., 8, 2000
|
|
1PL4
 
 | | Crystal Structure of human MnSOD Y166F mutant | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Fan, L, Tainer, J.A. | | Deposit date: | 2003-06-06 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Amino acid substitution at the dimeric interface of human manganese superoxide dismutase
J.Biol.Chem., 279, 2004
|
|
8CTH
 
 | | Cryo-EM structure of human METTL1-WDR4-tRNA(Phe) complex | | Descriptor: | Phe-tRNA, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(7)-)-methyltransferase, ... | | Authors: | Li, J, Wang, L, Fontana, P, Hunkeler, M, Roy-Burman, S.S, Wu, H, Fishcer, E.S, Gregory, R.I. | | Deposit date: | 2022-05-14 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
5XSN
 
 | | The catalytic domain of GdpP with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, MANGANESE (II) ION, Phosphodiesterase acting on cyclic dinucleotides | | Authors: | Wang, F, Gu, L. | | Deposit date: | 2017-06-14 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structural and biochemical characterization of the catalytic domains of GdpP reveals a unified hydrolysis mechanism for the DHH/DHHA1 phosphodiesterase
Biochem. J., 475, 2018
|
|
1G7Q
 
 | | CRYSTAL STRUCTURE OF MHC CLASS I H-2KB HEAVY CHAIN COMPLEXED WITH BETA-2 MICROGLOBULIN AND MUC1 VNTR PEPTIDE SAPDTRPA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-2 MICROGLOBULIN, ... | | Authors: | Apostolopoulos, V, Yu, M, Corper, A.L, Teyton, L, Pietersz, G.A, McKenzie, I.F, Wilson, I.A. | | Deposit date: | 2000-11-13 | | Release date: | 2002-07-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a non-canonical low-affinity peptide complexed with MHC class I: a new approach for vaccine design.
J.Mol.Biol., 318, 2002
|
|
8S8O
 
 | | Solution Structure of cAMP-dependent Protein Kinase RII-alpha Subunit Dimerization and Docking Domain Complex with Microtubule Associated Protein 2c (84-111) | | Descriptor: | Isoform 3 of Microtubule-associated protein 2, cAMP-dependent protein kinase type II-alpha regulatory subunit | | Authors: | Bartosik, V, Lanikova, A, Janackova, Z, Padrta, P, Zidek, L. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-25 | | Method: | SOLUTION NMR | | Cite: | Structural basis of binding the unique N-terminal domain of microtubule-associated protein 2c to proteins regulating kinases of signaling pathways.
J.Biol.Chem., 300, 2024
|
|
8CTI
 
 | | Cryo-EM structure of human METTL1-WDR4-tRNA(Val) complex | | Descriptor: | tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4, tRNA-Val-TAC-2-1 | | Authors: | Li, J, Wang, L, Fontana, P, Hunkeler, M, Roy-Burman, S.S, Wu, H, Fischer, E.S, Gregory, R.I. | | Deposit date: | 2022-05-14 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
6G8K
 
 | | 14-3-3sigma in complex with a S131beta3S mutated YAP pS127 phosphopeptide | | Descriptor: | 14-3-3 protein sigma, ACE-ARG-ALA-HIS-SEP-SER-PRO-ALA-BSE-LEU-GLN, CHLORIDE ION, ... | | Authors: | Andrei, S.A, Thijssen, V, Brunsveld, L, Ottmann, C, Milroy, L.G. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A study on the effect of synthetic alpha-to-beta3-amino acid mutations on the binding of phosphopeptides to 14-3-3 proteins.
Chem.Commun.(Camb.), 55, 2019
|
|
3UU8
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Loop2-24' mutant reduced in solution | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Nury, H, Corringer, P.J, Delarue, M. | | Deposit date: | 2011-11-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | A locally closed conformation of a bacterial pentameric proton-gated ion channel.
Nat.Struct.Mol.Biol., 19, 2012
|
|
8RW1
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-02-02 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
5M3W
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-1,2-dideoxymannose and alpha-1,2-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-10-17 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
3U9S
 
 | | Crystal structure of P. aeruginosa 3-methylcrotonyl-CoA carboxylase (MCC) 750 kD holoenzyme, CoA complex | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Methylcrotonyl-CoA carboxylase, ... | | Authors: | Huang, C.S, Tong, L. | | Deposit date: | 2011-10-19 | | Release date: | 2011-12-14 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | An unanticipated architecture of the 750-kDa {alpha}6{beta}6 holoenzyme of 3-methylcrotonyl-CoA carboxylase
Nature, 481, 2012
|
|
8D8I
 
 | | Crystal structure of Reverb alpha in complex with synthetic agonist | | Descriptor: | (4S)-6-[([1,1'-biphenyl]-2-yl)oxy]-3-chloro[1,2,4]triazolo[4,3-b]pyridazine, Nuclear receptor corepressor 1, Nuclear receptor subfamily 1 group D member 1 | | Authors: | Ronin, C, Ciesielski, F, Hegazy, L, Burris, P.T. | | Deposit date: | 2022-06-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural basis of synthetic agonist activation of the nuclear receptor REV-ERB.
Nat Commun, 13, 2022
|
|
6GC2
 
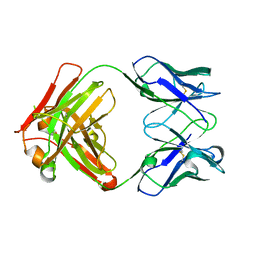 | | AbLIFT: Antibody stability and affinity optimization by computational design of the variable light-heavy chain interface | | Descriptor: | Heavy chain, Light Chain | | Authors: | Warszawski, S, Katz, A, Khmelnitsky, L, Ben Nissan, G, Javitt, G, Dym, O, Unger, T, Knop, O, Diskin, R, Albeck, S, Fass, D, Sharon, M, Fleishman, S.J. | | Deposit date: | 2018-04-17 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Optimizing antibody affinity and stability by the automated design of the variable light-heavy chain interfaces.
Plos Comput.Biol., 15, 2019
|
|
3TUS
 
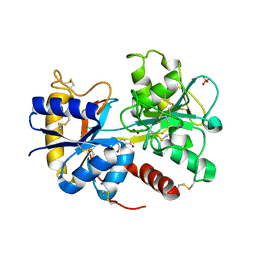 | | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Meta-hydroxy benzoic acid at 2.5 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-HYDROXYBENZOIC ACID, ... | | Authors: | Shukla, P.K, Gautam, L, Singh, A, Kaushik, S, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-09-18 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of C-lobe of Bovine lactoferrin Complexed with Meta-hydroxy benzoic acid at 2.5 A Resolution
To be Published
|
|
1Q0W
 
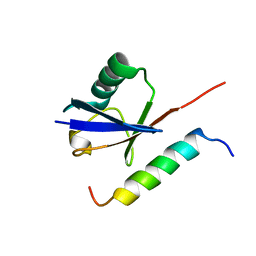 | | Solution structure of Vps27 amino-terminal UIM-ubiquitin complex | | Descriptor: | Ubiquitin, Vacuolar protein sorting-associated protein VPS27 | | Authors: | Swanson, K.A, Kang, R.S, Stamenova, S.D, Hicke, L, Radhakrishnan, I. | | Deposit date: | 2003-07-17 | | Release date: | 2003-10-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Vps27 UIM-Ubiquitin Complex Important for Endosomal Sorting and Receptor Downregulation
Embo J., 22, 2003
|
|
5XXB
 
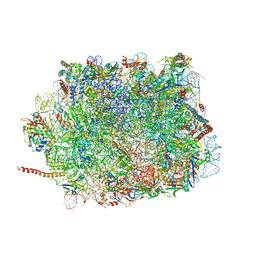 | | Large subunit of Toxoplasma gondii ribosome | | Descriptor: | 25S RNA, 5.8S RNA, 5S RNA, ... | | Authors: | Li, Z, Guo, Q, Zheng, L, Ji, Y, Xie, Y, Lai, D, Lun, Z, Suo, X, Gao, N. | | Deposit date: | 2017-07-03 | | Release date: | 2017-08-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cryo-EM structures of the 80S ribosomes from human parasites Trichomonas vaginalis and Toxoplasma gondii
Cell Res., 27, 2017
|
|
8S8E
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-3.1) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
8S8F
 
 | | Structure of a yeast 48S-AUC preinitiation complex in closed conformation (model py48S-AUC-3.2) | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein L41-A, 40S ribosomal protein S12, ... | | Authors: | Villamayor-Belinchon, L, Sharma, P, Llacer, J.L, Hussain, T. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Structural basis of AUC codon discrimination during translation initiation in yeast.
Nucleic Acids Res., 52, 2024
|
|
5XB7
 
 | | GH42 alpha-L-arabinopyranosidase from Bifidobacterium animalis subsp. lactis Bl-04 | | Descriptor: | Beta-galactosidase, GLYCEROL, SULFATE ION | | Authors: | Viborg, A.H, Katayama, T, Arakawa, T, Abou Hachem, M, Lo Leggio, L, Kitaoka, M, Svensson, B, Fushinobu, S. | | Deposit date: | 2017-03-16 | | Release date: | 2017-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of alpha-l-arabinopyranosidases from human gut microbiome expands the diversity within glycoside hydrolase family 42.
J. Biol. Chem., 292, 2017
|
|
