3I12
 
 | | The crystal structure of the D-alanyl-alanine synthetase A from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-alanine-D-alanine ligase A | | Authors: | Zhang, R, Maltseva, N, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-25 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structure of the D-alanyl-alanine synthetase A from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2
To be Published
|
|
4QI9
 
 | | Crystal structure of dihydrofolate reductase from Yersinia pestis complexed with methotrexate | | Descriptor: | Dihydrofolate reductase, METHOTREXATE | | Authors: | Maltseva, N, Kim, Y, Makowska-Grzyska, M, Mulligan, R, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-05-30 | | Release date: | 2014-07-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | Crystal structure of dihydrofolate reductase from Yersinia pestis complexed with methotrexate
To be Published
|
|
3LAY
 
 | | Alpha-Helical barrel formed by the decamer of the zinc resistance-associated protein (STM4172) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | Zinc resistance-associated protein | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Winsor, J, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-07 | | Release date: | 2010-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Alpha-Helical barrel formed by the decamer of the zinc resistance-associated protein (STM4172) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2
To be Published
|
|
4QJ1
 
 | | Co-crystal structure of the catalytic domain of the inosine monophosphate dehydrogenase from Cryptosporidium parvum with inhibitor N109 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, FORMIC ACID, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-03 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.415 Å) | | Cite: | Co-crystal structure of the catalytic domain of the inosine monophosphate dehydrogenase from Cryptosporidium parvum with inhibitor N109
To be Published
|
|
4QN2
 
 | | 2.6 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) G234S mutant from Staphylococcus aureus (IDP00699) in complex with NAD+ and BME-free Cys289 | | Descriptor: | ACETATE ION, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional analysis of betaine aldehyde dehydrogenase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4DYU
 
 | | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10 | | Descriptor: | DNA protection during starvation protein, SULFATE ION, ZINC ION | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-02-29 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The crystal structure of DNA starvation/stationary phase protection protein Dps from Yersinia pestis KIM 10
To be Published
|
|
4E16
 
 | | Precorrin-4 C(11)-methyltransferase from Clostridium difficile | | Descriptor: | precorrin-4 C(11)-methyltransferase | | Authors: | Osipiuk, J, Nocek, B, Makowska-Grzyska, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-05 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Precorrin-4 C(11)-methyltransferase from Clostridium difficile
To be Published
|
|
6DAU
 
 | | Crystal structure of E33Q and E41Q mutant forms of the spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae | | Descriptor: | GLYCEROL, Spermidine N1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Beahan, A, Kulyavtsev, P, Tan, L, Tran, D, Kuhn, M.L, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of E33Q and E41Q mutant forms of the spermidine/spermine N-acetyltransferase SpeG from Vibrio cholerae.
To be Published
|
|
3LNO
 
 | | Crystal Structure of Domain of Unknown Function DUF59 from Bacillus anthracis | | Descriptor: | Putative uncharacterized protein | | Authors: | Kim, Y, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Domain of Unknown Function DUF59 Bacillus anthracis
To be Published
|
|
4R7Q
 
 | | The structure of a sensor domain of a histidine kinase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | ACETATE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Sensor Domain of Histidine Kinase VxrA of Vibrio cholerae - A Hairpin-swapped Dimer and its Conformational Change.
J.Bacteriol., 203, 2021
|
|
4R9M
 
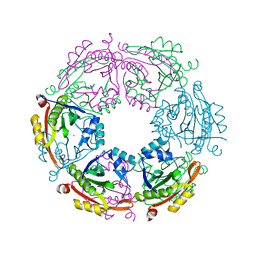 | | Crystal structure of spermidine N-acetyltransferase from Escherichia coli | | Descriptor: | MAGNESIUM ION, Spermidine N(1)-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Shuvalova, L, Grimshaw, S, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Analysis of crystalline and solution states of ligand-free spermidine N-acetyltransferase (SpeG) from Escherichia coli.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6C5C
 
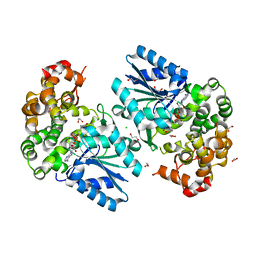 | | Crystal structure of the 3-dehydroquinate synthase (DHQS) domain of Aro1 from Candida albicans SC5314 in complex with NADH | | Descriptor: | 1,2-ETHANEDIOL, 3-dehydroquinate synthase, CHLORIDE ION, ... | | Authors: | Michalska, K, Evdokimova, E, Di Leo, R, Stogios, P.J, Savchenko, A, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-16 | | Release date: | 2018-01-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular analysis and essentiality of Aro1 shikimate biosynthesis multi-enzyme in Candida albicans.
Life Sci Alliance, 5, 2022
|
|
4QVT
 
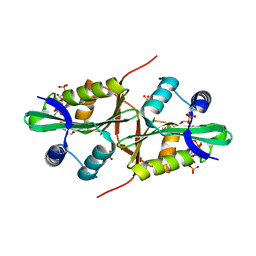 | | Crystal structure of predicted N-acyltransferase (ypeA) in complex with acetyl-CoA from Escherichia coli | | Descriptor: | ACETYL COENZYME *A, Acetyltransferase YpeA, SULFATE ION | | Authors: | Filippova, E.V, Minasov, G, Winsor, G, Dubrovska, I, Shuvalova, L, Wolfe, A.J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-15 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Crystal structure of predicted N-acyltransferase (ypeA) in complex with acetyl-CoA from Escherichia coli
To be Published
|
|
3LUS
 
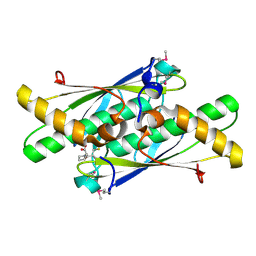 | | Crystal structure of a putative organic hydroperoxide resistance protein with molecule of captopril bound in one of the active sites from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | L-CAPTOPRIL, Organic hydroperoxide resistance protein | | Authors: | Nocek, B, Maltseva, N, Makowska-Grzyska, M, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-18 | | Release date: | 2010-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of a putative organic hydroperoxide resistance protein with molecule of captopril bound in one of the active sites from Vibrio cholerae O1 biovar eltor str. N16961
TO BE PUBLISHED
|
|
4QUS
 
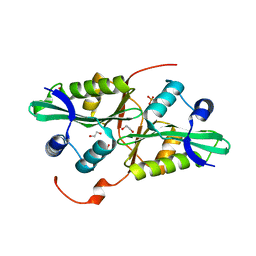 | | 1.28 Angstrom resolution crystal structure of predicted acyltransferase with acyl-CoA N-acyltransferase domain (ypeA) from Escherichia coli str. K-12 substr. MG1655 | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase YpeA, PHOSPHATE ION | | Authors: | Halavaty, A.S, Filippova, E.V, Minasov, G, Flores, K.J, Dubrovska, I, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-07-11 | | Release date: | 2014-07-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | 1.28 Angstrom resolution crystal structure of predicted acyltransferase with acyl-CoA N-acyltransferase domain (ypeA) from Escherichia coli str. K-12 substr. MG1655
To be Published
|
|
4QQ3
 
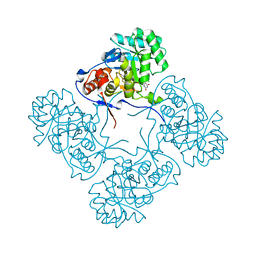 | | Inosine 5'-monophosphate dehydrogenase from Vibrio cholerae, deletion mutant, in complex with XMP | | Descriptor: | CHLORIDE ION, Inosine-5'-monophosphate dehydrogenase, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Osipiuk, J, Maltseva, N, Makowska-Grzyska, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-06-26 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Inosine 5'-monophosphate dehydrogenase from vibrio cholerae, deletion mutant, in complex with xmp
To be Published
|
|
4FEU
 
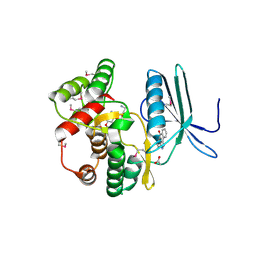 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor anthrapyrazolone SP600125 | | Descriptor: | 2,6-DIHYDROANTHRA/1,9-CD/PYRAZOL-6-ONE, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
3M7W
 
 | | Crystal Structure of Type I 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2 in Covalent Complex with Dehydroquinate | | Descriptor: | 1,3,4-TRIHYDROXY-5-OXO-CYCLOHEXANECARBOXYLIC ACID, 3-dehydroquinate dehydratase, GLYCEROL | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-17 | | Release date: | 2010-04-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the mechanism of type I dehydroquinate dehydratases from structures of reaction intermediates.
J.Biol.Chem., 286, 2011
|
|
6DWE
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - aminoacrylate- and BRD0059-bound form | | Descriptor: | (2R,3S,4R)-3-(2',6'-difluoro-4'-methyl[1,1'-biphenyl]-4-yl)-4-(fluoromethyl)azetidine-2-carbonitrile, 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, ... | | Authors: | Chang, C, Michalska, K, Maltseva, N.I, Jedrzejczak, R, McCarren, P, Nag, P.P, Joachimiak, A, Satchell, K, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-06-26 | | Release date: | 2018-07-11 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Crystal structure of tryptophan synthase from M. tuberculosis - closed form with BRD6309 bound
To be Published
|
|
7JIR
 
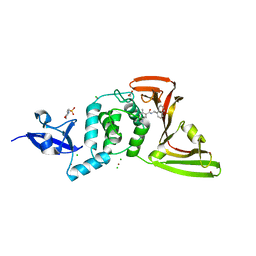 | | The crystal structure of Papain-Like Protease of SARS CoV-2 , C111S mutant, in complex with PLP_Snyder457 inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl]benzamide, ACETATE ION, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Lisnyak, V, Maki, S, Taylor, C, Zhang, Y, Zhou, Z, Azizi, S.A, Jones, K, Kathayat, R, Snyder, S.A, Dickinson, B.C, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors.
Nat Commun, 12, 2021
|
|
4FEW
 
 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor pyrazolopyrimidine PP2 | | Descriptor: | 1-TERT-BUTYL-3-(4-CHLORO-PHENYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
5ULB
 
 | |
3MSU
 
 | | Crystal Structure of Citrate Synthase from Francisella tularensis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETIC ACID, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-04-29 | | Release date: | 2010-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.843 Å) | | Cite: | Crystal Structure of Citrate Synthase from Francisella tularensis
To be Published
|
|
4Q7G
 
 | | 1.7 Angstrom Crystal Structure of leukotoxin LukD from Staphylococcus aureus. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Leucotoxin LukDv | | Authors: | Minasov, G, Nocadello, S, Shuvalova, L, Shatsman, S, Kwon, K, Bagnoli, F, Falugi, F, Bottomley, M, Grandi, G, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-04-24 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the components of the Staphylococcus aureus leukotoxin ED.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
7MBL
 
 | | Crystal structure of Equine Serum Albumin in complex with Cobalt (II) | | Descriptor: | COBALT (II) ION, SULFATE ION, Serum albumin | | Authors: | Shabalin, I.G, Czub, M.P, Handing, K.B, Cymborowski, M.T, Grabowski, M, Cooper, D.R, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-03-31 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical characterisation of Co 2+ -binding sites on serum albumins and their interplay with fatty acids.
Chem Sci, 14, 2023
|
|
