4O9U
 
 | | Mechanism of transhydrogenase coupling proton translocation and hydride transfer | | Descriptor: | NAD(P) transhydrogenase subunit alpha 2, NAD(P) transhydrogenase subunit beta, NAD/NADP transhydrogenase alpha subunit 1, ... | | Authors: | Leung, J.H, Yamaguchi, M, Moeller, A, Schurig-Briccio, L.A, Gennis, R.B, Potter, C.S, Carragher, B, Stout, C.D. | | Deposit date: | 2014-01-02 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (6.926 Å) | | Cite: | Structural biology. Division of labor in transhydrogenase by alternating proton translocation and hydride transfer.
Science, 347, 2015
|
|
6QQI
 
 | |
4U39
 
 | | Crystal Structure of FtsZ:MciZ Complex from Bacillus subtilis | | Descriptor: | Cell division factor, Cell division protein FtsZ, PHOSPHATE ION | | Authors: | Bisson-Filho, A.W, Discola, K.F, Castellen, P, Blasios, V, Martins, A, Sforca, M.L, Garcia, W, Zeri, A.C, Erickson, H.P, Dessen, A, Gueiros-Filho, F.J. | | Deposit date: | 2014-07-19 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Crystal Structure of FtsZ:MciZ Complex from Bacillus subtilis
To be Published
|
|
1N2T
 
 | | C-DES Mutant K223A with GLY Covalenty Linked to the PLP-cofactor | | Descriptor: | GLYCINE, L-cysteine/cystine lyase C-DES, POTASSIUM ION, ... | | Authors: | Kaiser, J.T, Bruno, S, Clausen, T, Huber, R, Schiaretti, F, Mozzarelli, A, Kessler, D. | | Deposit date: | 2002-10-24 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Snapshots of the Cystine Lyase "C-DES" during Catalysis: Studies in Solution and in the Crystalline State
J.Biol.Chem., 278, 2003
|
|
1MR5
 
 | | Orthorhombic form of Trypanosoma cruzi trans-sialidase | | Descriptor: | trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-18 | | Release date: | 2003-03-18 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
1MRX
 
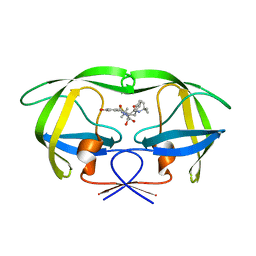 | | Structure of HIV protease (Mutant Q7K L33I L63I V82F I84V ) complexed with KNI-577 | | Descriptor: | (4R)-N-tert-butyl-3-{(2S,3S)-2-hydroxy-3-[(3-hydroxy-2-methylbenzoyl)amino]-4-phenylbutanoyl}-5,5-dimethyl-1,3-thiazoli dine-4-carboxamide, POL polyprotein | | Authors: | Vega, S, Kang, L.-W, Velazquez-Campoy, A, Kiso, Y, Amzel, L.M, Freire, E. | | Deposit date: | 2002-09-18 | | Release date: | 2003-11-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and thermodynamic escape mechanism from a drug resistant mutation of the HIV-1 protease.
Proteins, 55, 2004
|
|
1MS8
 
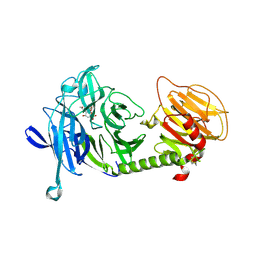 | | Triclinic form of Trypanosoma cruzi trans-sialidase, in complex with 3-deoxy-2,3-dehydro-N-acetylneuraminic acid (DANA) | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, trans-sialidase | | Authors: | Buschiazzo, A, Amaya, M.F, Cremona, M.L, Frasch, A.C, Alzari, P.M. | | Deposit date: | 2002-09-19 | | Release date: | 2003-03-25 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure and mode of action of trans-sialidase, a key enzyme in Trypanosoma cruzi pathogenesis
Mol.Cell, 10, 2002
|
|
4OD0
 
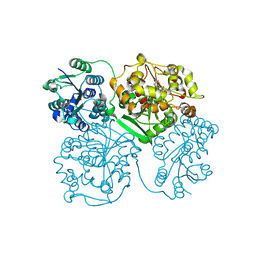 | | Crystal structure of human soluble epoxide hydrolase complexed with 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea | | Descriptor: | 1-(1-propanoylpiperidin-4-yl)-3-[4-(trifluoromethoxy)phenyl]urea, Bifunctional epoxide hydrolase 2, MAGNESIUM ION, ... | | Authors: | Lee, K.S.S, Liu, J, Wagner, K.M, Pakhomova, S, Dong, H, Morisseau, C, Fu, S.H, Yang, J, Wang, P, Ulu, A, Mate, C, Nguyen, L, Wullf, H, Eldin, M.L, Mara, A.A, Newcomer, M.E, Zeldin, D.C, Hammock, B.D. | | Deposit date: | 2014-01-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Optimized inhibitors of soluble epoxide hydrolase improve in vitro target residence time and in vivo efficacy.
J.Med.Chem., 57, 2014
|
|
4O6Z
 
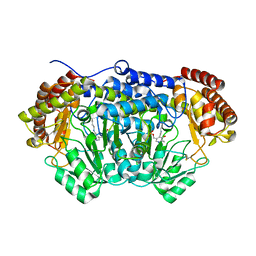 | | Crystal structure of serine hydroxymethyltransferase with covalently bound PLP Schiff-base from Plasmodium falciparum | | Descriptor: | PHOSPHATE ION, PYRIDOXAL-5'-PHOSPHATE, Serine hydroxymethyltransferase | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U. | | Deposit date: | 2013-12-24 | | Release date: | 2014-06-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | The structure of Plasmodium falciparum serine hydroxymethyltransferase reveals a novel redox switch that regulates its activities.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1MUZ
 
 | |
4OEM
 
 | | Crystal structure of Cathepsin C in complex with dipeptide substrates | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Zhao, B, Smallwood, A, Concha, N. | | Deposit date: | 2014-01-13 | | Release date: | 2015-03-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The amino-acid substituents of dipeptide substrates of cathepsin C can determine the rate-limiting steps of catalysis.
Biochemistry, 51, 2012
|
|
4OAK
 
 | | Crystal structure of vancomycin resistance D,D-dipeptidase/D,D-pentapeptidase VanXYc D59S mutant in complex with D-Alanine-D-Alanine and copper (II) | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Meziane-Cherif, D, Di Leo, R, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-01-04 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the evolution of vancomycin resistance D,D-peptidases.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1N06
 
 | | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, PUTATIVE riboflavin kinase | | Authors: | Bauer, S, Kemter, K, Bacher, A, Huber, R, Fischer, M, Steinbacher, S. | | Deposit date: | 2002-10-11 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Schizosaccharomyces pombe Riboflavin Kinase Reveals a Novel ATP and Riboflavin Binding Fold
J.Mol.Biol., 326, 2003
|
|
4U7D
 
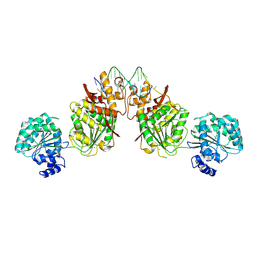 | | Structure of human RECQ-like helicase in complex with an oligonucleotide | | Descriptor: | ATP-dependent DNA helicase Q1, DNA oligonucleotide, ZINC ION | | Authors: | Pike, A.C.W, Zhang, Y, Schnecke, C, Cooper, C.D.O, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-07-30 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Human RECQ1 helicase-driven DNA unwinding, annealing, and branch migration: Insights from DNA complex structures.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1MYZ
 
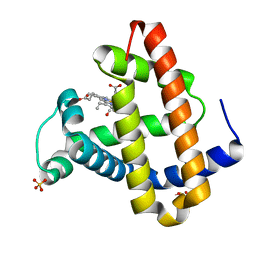 | | CO COMPLEX OF MYOGLOBIN MB-YQR AT RT SOLVED FROM LAUE DATA. | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bourgeois, D, Vallone, B, Schotte, F, Arcovito, A, Miele, A.E, Sciara, G, Wulff, M, Anfinrud, P, Brunori, M. | | Deposit date: | 2002-10-04 | | Release date: | 2003-08-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Complex landscape of protein
structural dynamics unveiled by
nanosecond Laue crystallography.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1MZI
 
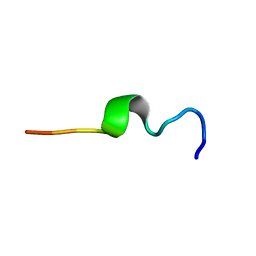 | | Solution ensemble structures of HIV-1 gp41 2F5 mAb epitope | | Descriptor: | 2F5 epitope of HIV-1 gp41 fusion protein | | Authors: | Barbato, G, Bianchi, E, Ingallinella, P, Hurni, W.H, Miller, M.D, Ciliberto, G, Cortese, R, Bazzo, R, Shiver, J.W, Pessi, A. | | Deposit date: | 2002-10-08 | | Release date: | 2003-09-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of the epitope of the anti-HIV antibody 2F5 sheds light into its mechanism of neutralization and HIV fusion.
J.Mol.Biol., 330, 2003
|
|
1N1O
 
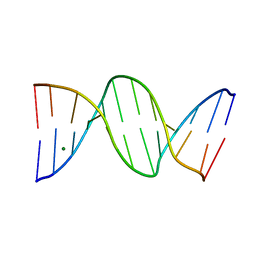 | | Crystal Structure of a B-form DNA Duplex Containing (L)-alpha-threofuranosyl (3'-2') Nucleosides: A Four-Carbon Sugar is Easily Accommodated into the Backbone of DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*(TFT)P*TP*CP*GP*CP*G)-3', MAGNESIUM ION | | Authors: | Wilds, C.J, Wawrzak, Z, Krishnamurthy, R, Eschenmoser, A, Egli, M. | | Deposit date: | 2002-10-18 | | Release date: | 2002-11-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structure of a B-Form DNA Duplex Containing (L)-alpha-Threofuranosyl (3'-->2') Nucleosides: A
Four-Carbon Sugar Is Easily Accommodated into the Backbone of DNA
J.Am.Chem.Soc., 124, 2002
|
|
1N4V
 
 | |
4OMN
 
 | |
4OMQ
 
 | |
1N26
 
 | | Crystal Structure of the extra-cellular domains of Human Interleukin-6 Receptor alpha chain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Varghese, J.N, Moritz, R.L, Lou, M.-Z, van Donkelaar, A, Ji, H, Ivancic, N, Branson, K.M, Hall, N.E, Simpson, R.J. | | Deposit date: | 2002-10-22 | | Release date: | 2002-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the extracellular domains of the human interleukin-6 receptor alpha-chain.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
4OI5
 
 | | Glycerol-free structure of thermolysin in complex with ubtln58 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, P-((((benzyloxy)carbonyl)amino)methyl)-N-((S)-1-((3,3-dimethylbutyl)amino)-4-methyl-1-oxopentan-2-yl)phosphonamidic acid, ... | | Authors: | Krimmer, S.G, Heine, A, Klebe, G. | | Deposit date: | 2014-01-18 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Methyl, Ethyl, Propyl, Butyl: Futile But Not for Water, as the Correlation of Structure and Thermodynamic Signature Shows in a Congeneric Series of Thermolysin Inhibitors.
Chemmedchem, 9, 2014
|
|
4UCW
 
 | | Structure of the T18V small subunit mutant of D. fructosovorans NiFe- hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Abou-Hamdan, A, Ceccaldi, P, Lebrette, H, Guttierez-Sanz, O, Richaud, P, Cournac, L, Guigliarelli, B, deLacey, A.L, Leger, C, Volbeda, A, Burlat, B, Dementin, S. | | Deposit date: | 2014-12-04 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Threonine Stabilizes the Nic and Nir Catalytic Intermediates of [Nife]-Hydrogenase.
J.Biol.Chem., 290, 2015
|
|
7RDF
 
 | | Crystal structure of Pseudomonas aeruginosa D-Arginine Dehydrogenase Y249F co-crystallized in the presence of D-arginine | | Descriptor: | 6-HYDROXY-FLAVIN-ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, FAD-dependent catabolic D-arginine dehydrogenase DauA, ... | | Authors: | Reis, R.A.G, Iyer, A, Agniswamy, A, Weber, I.T, Gadda, G. | | Deposit date: | 2021-07-09 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Discovery of a new flavin N5-adduct in a tyrosine to phenylalanine variant of d-Arginine dehydrogenase.
Arch.Biochem.Biophys., 715, 2021
|
|
4UCX
 
 | | Structure of the T18G small subunit mutant of D. fructosovorans NiFe- hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Abou-Hamdan, A, Ceccaldi, P, Lebrette, H, Guttierez-Sanz, O, Richaud, P, Cournac, L, Guigliarelli, B, deLacey, A.L, Leger, C, Volbeda, A, Burlat, B, Dementin, S. | | Deposit date: | 2014-12-05 | | Release date: | 2015-02-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A threonine stabilizes the NiC and NiR catalytic intermediates of [NiFe]-hydrogenase.
J. Biol. Chem., 290, 2015
|
|
