5D23
 
 | |
4NWL
 
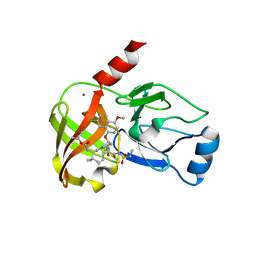 | | Crystal structure of hepatis c virus protease (ns3) complexed with bms-650032 aka n-(tert-butoxycarbonyl)-3-me thyl-l-valyl-(4r)-4-((7-chloro-4-methoxy-1-isoquinolinyl)o xy)-n-((1r,2s)-1-((cyclopropylsulfonyl)carbamoyl)-2-vinylc yclopropyl)-l-prolinamide | | Descriptor: | HCV NS3 1a Protease, N-(tert-butoxycarbonyl)-3-methyl-L-valyl-(4R)-4-[(7-chloro-4-methoxyisoquinolin-1-yl)oxy]-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-L-prolinamide, ZINC ION | | Authors: | Muckelbauer, J.K, Klei, H.E. | | Deposit date: | 2013-12-06 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Early Clinical Evaluation of BMS-605339, a Potent and Orally Efficacious Tripeptidic Acylsulfonamide NS3 Protease Inhibitor for the Treatment of Hepatitis C Virus Infection.
J.Med.Chem., 57, 2014
|
|
4FJZ
 
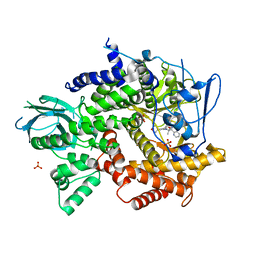 | | Crystal structure of PI3K-gamma in complex with pyrrolo-pyridine inhibitor 63 | | Descriptor: | 1'-[7-fluoro-3-methyl-2-(pyridin-2-yl)quinolin-4-yl]-6'-(morpholin-4-yl)-1',2,2',3,5,6-hexahydrospiro[pyran-4,3'-pyrrolo[3,2-b]pyridine], Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-06-12 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery and in Vivo Evaluation of Dual PI3K-beta/delta inhibitors
J.Med.Chem., 55, 2012
|
|
4FJY
 
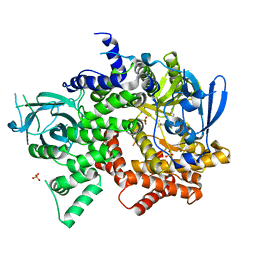 | | Crystal structure of PI3K-gamma in complex with quinoline-indoline inhibitor 24f | | Descriptor: | 4-[3,3-dimethyl-6-(morpholin-4-yl)-2,3-dihydro-1H-indol-1-yl]-7-fluoro-3-methyl-2-(pyridin-3-yl)quinoline, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-06-12 | | Release date: | 2012-10-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and in Vivo Evaluation of Dual PI3K-beta/delta inhibitors
J.Med.Chem., 55, 2012
|
|
7XD2
 
 | |
6QZD
 
 | | HLA-DR1 with SGP Influenza Matrix Peptide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Greenshields-Watson, A, Rizkallah, P.J. | | Deposit date: | 2019-03-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | CD4+T Cells Recognize Conserved Influenza A Epitopes through Shared Patterns of V-Gene Usage and Complementary Biochemical Features.
Cell Rep, 32, 2020
|
|
6QZA
 
 | | HLA-DR1 with GMF Influenza PB1 Peptide | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, DRB1-1 beta chain, ... | | Authors: | Greenshields-Watson, A, Rizkallah, P.J. | | Deposit date: | 2019-03-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | CD4+T Cells Recognize Conserved Influenza A Epitopes through Shared Patterns of V-Gene Usage and Complementary Biochemical Features.
Cell Rep, 32, 2020
|
|
6R0E
 
 | |
8XGT
 
 | |
8XFV
 
 | |
8XGA
 
 | |
8XGB
 
 | |
8XGY
 
 | |
2FVD
 
 | | Cyclin Dependent Kinase 2 (CDK2) with diaminopyrimidine inhibitor | | Descriptor: | (4-AMINO-2-{[1-(METHYLSULFONYL)PIPERIDIN-4-YL]AMINO}PYRIMIDIN-5-YL)(2,3-DIFLUORO-6-METHOXYPHENYL)METHANONE, Cell division protein kinase 2 | | Authors: | Crowther, R.L, Lukacs, C.M, Kammlott, R.U. | | Deposit date: | 2006-01-30 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery of [4-Amino-2-(1-methanesulfonylpiperidin-4-ylamino)pyrimidin-5-yl](2,3-difluoro-6- methoxyphenyl)methanone (R547), a potent and selective cyclin-dependent kinase inhibitor with significant in vivo antitumor activity.
J.Med.Chem., 49, 2006
|
|
5VSN
 
 | |
7BPU
 
 | |
8KHD
 
 | | The interface structure of Omicron RBD binding to 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817, Light chain of 5817, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
8KHC
 
 | | SARS-CoV-2 Omicron spike in complex with 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817 Fab, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
5KCJ
 
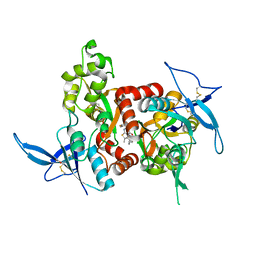 | | Structure of the human GluN1/GluN2A LBD in complex with GNE6901 | | Descriptor: | 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-06-06 | | Release date: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Positive Allosteric Modulators of GluN2A-Containing NMDARs with Distinct Modes of Action and Impacts on Circuit Function.
Neuron, 89, 2016
|
|
4FA7
 
 | |
7Y8G
 
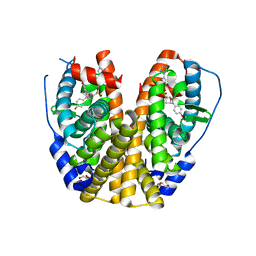 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with an Inhibitor 30a and GRIP Peptide | | Descriptor: | DI(HYDROXYETHYL)ETHER, Estrogen receptor, Grip peptide, ... | | Authors: | Min, J, Hu, H.B, Yang, Y, Dong, C.E, Zhou, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-23 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure-guided identification of novel dual-targeting estrogen receptor alpha degraders with aromatase inhibitory activity for the treatment of endocrine-resistant breast cancer.
Eur.J.Med.Chem., 253, 2023
|
|
7Y8F
 
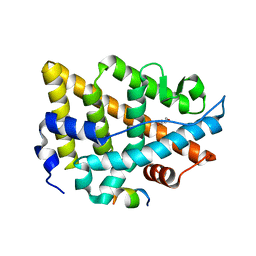 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with an Inhibitor 30o and GRIP Peptide | | Descriptor: | DI(HYDROXYETHYL)ETHER, Estrogen receptor, Grip peptide, ... | | Authors: | Min, J, Hu, H.B, Yang, Y, Dong, C.E, Zhou, H.B, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-23 | | Release date: | 2023-04-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structure-guided identification of novel dual-targeting estrogen receptor alpha degraders with aromatase inhibitory activity for the treatment of endocrine-resistant breast cancer.
Eur.J.Med.Chem., 253, 2023
|
|
4XKX
 
 | | Crystal structure of BACE1 in complex with 2-aminooxazoline 3-azaxanthene inhibitor 28 | | Descriptor: | (5S)-7-(2-fluoropyridin-3-yl)-3-(2-fluoropyridin-4-yl)spiro[chromeno[2,3-c]pyridine-5,4'-[1,3]oxazol]-2'-amine, Beta-secretase 1, GLYCEROL, ... | | Authors: | Whittington, D.A, Long, A.M. | | Deposit date: | 2015-01-12 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of 2-aminooxazoline 3-azaxanthenes as orally efficacious beta-secretase inhibitors for the potential treatment of Alzheimer's disease.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
9F9M
 
 | | Crystal structure of MUS81-EME1 bound by compound 21. | | Descriptor: | 5-oxidanyl-4-oxidanylidene-1-(4-piperazin-1-ylphenyl)pyridine-3-carboxylic acid, Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81, ... | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-08 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.469 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors
Acs Med.Chem.Lett., 2024
|
|
9F98
 
 | | Crystal structure of MUS81-EME1, apo form. | | Descriptor: | Crossover junction endonuclease EME1, Crossover junction endonuclease MUS81 | | Authors: | Collie, G.W. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Discovery of Novel MUS81 Inhibitors
Acs Med.Chem.Lett., 2024
|
|
