2WXK
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with INK666. | | Descriptor: | 3-(2-amino-1,3-benzothiazol-6-yl)-1-{[2-(4-methylpiperazin-1-yl)quinolin-3-yl]methyl}-1H-pyrazolo[3,4-d]pyrimidin-4-amine, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
2WXL
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with ZSTK474. | | Descriptor: | 2-(difluoromethyl)-1-(4,6-dimorpholin-4-yl-1,3,5-triazin-2-yl)-1H-benzimidazole, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
2WXF
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with PIK-39. | | Descriptor: | 2-((9H-PURIN-6-YLTHIO)METHYL)-5-CHLORO-3-(2-METHOXYPHENYL)QUINAZOLIN-4(3H)-ONE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
2WXO
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with AS5. | | Descriptor: | N-(3-{[(1Z)-3,5-DIMETHOXYCYCLOHEXA-2,4-DIEN-1-YLIDENE]AMINO}QUINOXALIN-2-YL)-4-FLUOROBENZENESULFONAMIDE, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
2WXJ
 
 | | The crystal structure of the murine class IA PI 3-kinase p110delta in complex with INK654. | | Descriptor: | N-[6-(4-amino-1-{[2-(4-methylpiperazin-1-yl)quinolin-3-yl]methyl}-1H-pyrazolo[3,4-d]pyrimidin-3-yl)-1,3-benzothiazol-2-yl]acetamide, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT DELTA ISOFORM | | Authors: | Berndt, A, Miller, S, Williams, O, Lee, D.D, Houseman, B.T, Pacold, J.I, Gorrec, F, Hon, W.-C, Liu, Y, Rommel, C, Gaillard, P, Ruckle, T, Schwarz, M.K, Shokat, K.M, Shaw, J.P, Williams, R.L. | | Deposit date: | 2009-11-09 | | Release date: | 2010-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The P110D Structure: Mechanisms for Selectivity and Potency of New Pi(3)K Inhibitors
Nat.Chem.Biol., 6, 2010
|
|
8RCK
 
 | | CryoEM structure of mTORC1 with a paediatric kidney cancer-associated 1455-EWED-1458 duplication in mTOR, Focused on one protomer copy. | | Descriptor: | Eukaryotic translation initiation factor 4E-binding protein 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Anandapadamanaban, M, Hay, I.M, Perisic, O, Williams, R.L. | | Deposit date: | 2023-12-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CryoEM structure of mTORC1 with a paediatric kidney cancer-associated 1455-EWED-1458 duplication in mTOR, Focused on one protomer copy.
To Be Published
|
|
8RCN
 
 | | CryoEM structure of mTORC1 with a paediatric kidney cancer-associated 1455-EWED-1458 duplication in mTOR, Focused region of mTOR and RAPTOR on one protomer copy. | | Descriptor: | Eukaryotic translation initiation factor 4E-binding protein 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Anandapadamanaban, M, Hay, I.M, Perisic, O, Williams, R.L. | | Deposit date: | 2023-12-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | CryoEM structure of mTORC1 with a paediatric kidney cancer-associated 1455-EWED-1458 duplication in mTOR, Focused region of mTOR and RAPTOR on one protomer copy.
To Be Published
|
|
2X6I
 
 | | THE CRYSTAL STRUCTURE OF THE DROSOPHILA CLASS III PI3-KINASE VPS34 IN COMPLEX WITH PIK-90 | | Descriptor: | N-(2,3-DIHYDRO-7,8-DIMETHOXYIMIDAZO[1,2-C] QUINAZOLIN-5-YL)NICOTINAMIDE, PHOSPHOTIDYLINOSITOL 3 KINASE 59F | | Authors: | Miller, S, Tavshanjian, B, Oleksy, A, Perisic, O, Houseman, B.T, Shokat, K.M, Williams, R.L. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Shaping Development of Autophagy Inhibitors with the Structure of the Lipid Kinase Vps34.
Science, 327, 2010
|
|
2X6H
 
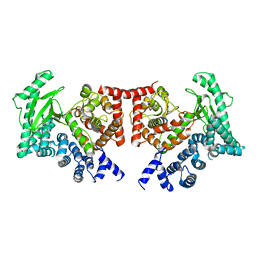 | | THE CRYSTAL STRUCTURE OF THE DROSOPHILA CLASS III PI3-KINASE VPS34 | | Descriptor: | PHOSPHOTIDYLINOSITOL 3 KINASE 59F | | Authors: | Miller, S, Tavshanjian, B, Oleksy, A, Perisic, O, Houseman, B.T, Shokat, K.M, Williams, R.L. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Shaping Development of Autophagy Inhibitors with the Structure of the Lipid Kinase Vps34.
Science, 327, 2010
|
|
2XVC
 
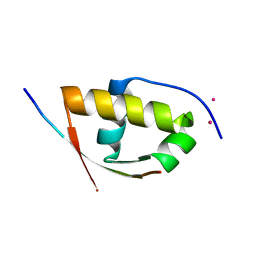 | | Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division | | Descriptor: | CADMIUM ION, CDVA, SSO0911, ... | | Authors: | Samson, R.Y, Obita, T, Hodgson, B, Shaw, M.K, Chong, P.L, Williams, R.L, Bell, S.D. | | Deposit date: | 2010-10-25 | | Release date: | 2011-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular and Structural Basis of Escrt-III Recruitment to Membranes During Archaeal Cell Division.
Mol.Cell, 41, 2011
|
|
2X6F
 
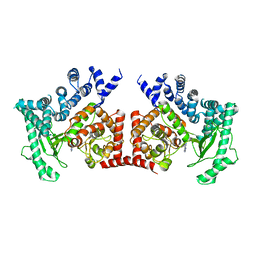 | | THE CRYSTAL STRUCTURE OF THE DROSOPHILA CLASS III PI3-KINASE VPS34 IN COMPLEX WITH 3-METHYLADENINE | | Descriptor: | 6-AMINO-3-METHYLPURINE, PHOSPHOTIDYLINOSITOL 3 KINASE 59F | | Authors: | Miller, S, Tavshanjian, B, Oleksy, A, Perisic, O, Houseman, B.T, Shokat, K.M, Williams, R.L. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Shaping Development of Autophagy Inhibitors with the Structure of the Lipid Kinase Vps34.
Science, 327, 2010
|
|
2X6J
 
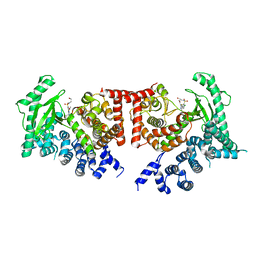 | | THE CRYSTAL STRUCTURE OF THE DROSOPHILA CLASS III PI3-KINASE VPS34 IN COMPLEX WITH PIK-93 | | Descriptor: | N-(5-(4-CHLORO-3-(2-HYDROXY-ETHYLSULFAMOYL)- PHENYLTHIAZOLE-2-YL)-ACETAMIDE, PHOSPHOTIDYLINOSITOL 3 KINASE 59F | | Authors: | Miller, S, Tavshanjian, B, Oleksy, A, Perisic, O, Houseman, B.T, Shokat, K.M, Williams, R.L. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Shaping Development of Autophagy Inhibitors with the Structure of the Lipid Kinase Vps34.
Science, 327, 2010
|
|
2X6K
 
 | | THE CRYSTAL STRUCTURE OF THE DROSOPHILA CLASS III PI3-KINASE VPS34 IN COMPLEX WITH PI-103 | | Descriptor: | 3-(4-MORPHOLIN-4-YLPYRIDO[3',2':4,5]FURO[3,2-D]PYRIMIDIN-2-YL)PHENOL, PHOSPHOTIDYLINOSITOL 3 KINASE 59F, SULFATE ION | | Authors: | Miller, S, Tavshanjian, B, Oleksy, A, Perisic, O, Houseman, B.T, Shokat, K.M, Williams, R.L. | | Deposit date: | 2010-02-17 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Shaping Development of Autophagy Inhibitors with the Structure of the Lipid Kinase Vps34.
Science, 327, 2010
|
|
2W2U
 
 | | STRUCTURAL INSIGHT INTO THE INTERACTION BETWEEN ARCHAEAL ESCRT-III AND AAA-ATPASE | | Descriptor: | CONSERVED ARCHAEAL PROTEIN, HYPOTHETICAL P60 KATANIN | | Authors: | Obita, T, Samson, R.Y, Perisic, O, Freund, S.M, Bell, S.D, Williams, R.L. | | Deposit date: | 2008-11-04 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Role for the Escrt System in Cell Division in Archaea.
Science, 322, 2008
|
|
2YMB
 
 | | Structures of MITD1 | | Descriptor: | CHARGED MULTIVESICULAR BODY PROTEIN 1A, MIT DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Hadders, M.A, Agromayor, M, Obita, T, Perisic, O, Caballe, A, Kloc, M, Lamers, M.H, Williams, R.L, Martin-Serrano, J. | | Deposit date: | 2012-10-08 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.404 Å) | | Cite: | Escrt-III Binding Protein Mitd1 is Involved in Cytokinesis and Has an Unanticipated Pld Fold that Binds Membranes.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2Y3A
 
 | | Crystal structure of p110beta in complex with icSH2 of p85beta and the drug GDC-0941 | | Descriptor: | 2-(1H-indazol-4-yl)-6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-yl-thieno[3,2-d]pyrimidine, PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT BETA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT BETA ISOFORM | | Authors: | Zhang, X, Vadas, O, Perisic, O, Williams, R.L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Lipid Kinase P110Beta-P85Beta Elucidates an Unusual Sh2-Domain-Mediated Inhibitory Mechanism.
Mol.Cell, 41, 2011
|
|
5NGB
 
 | |
1U5T
 
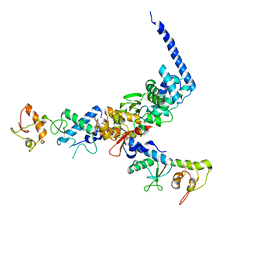 | | Structure of the ESCRT-II endosomal trafficking complex | | Descriptor: | Defective in vacuolar protein sorting; Vps36p, Hypothetical 23.6 kDa protein in YUH1-URA8 intergenic region, appears to be functionally related to SNF7; Snf8p | | Authors: | Hierro, A, Sun, J, Rusnak, A.S, Kim, J, Prag, G, Emr, S.D, Hurley, J.H. | | Deposit date: | 2004-07-28 | | Release date: | 2004-09-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of ESCRT-II endosomal trafficking complex
Nature, 431, 2004
|
|
1QAS
 
 | |
1QAT
 
 | |
5KC1
 
 | | Structure of the C-terminal dimerization domain of Atg38 | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Autophagy-related protein 38, ... | | Authors: | Ohashi, Y, Soler, N, Garcia-Ortegon, M, Zhang, L, Perisic, O, Masson, G.R, Johnson, C.M, Williams, R.J. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
7Z74
 
 | | PI3KC2a core in complex with PITCOIN2 | | Descriptor: | 1,2-ETHANEDIOL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, ~{N}-[4-(3-hydroxyphenyl)-1,3-thiazol-2-yl]-2-[4-oxidanylidene-3-(2-phenylethyl)pteridin-2-yl]sulfanyl-ethanamide | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2022-03-15 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of selective inhibitors of phosphatidylinositol 3-kinase C2 alpha.
Nat.Chem.Biol., 19, 2023
|
|
7Z75
 
 | | PI3KC2a core in complex with PITCOIN3 | | Descriptor: | 1,2-ETHANEDIOL, Phosphatidylinositol 4-phosphate 3-kinase C2 domain-containing subunit alpha, SULFATE ION, ... | | Authors: | Lo, W.T, Roske, Y, Daumke, O, Haucke, V. | | Deposit date: | 2022-03-15 | | Release date: | 2022-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Development of selective inhibitors of phosphatidylinositol 3-kinase C2 alpha.
Nat.Chem.Biol., 19, 2023
|
|
6GY0
 
 | | mPI3Kd IN COMPLEX WITH AZ3 | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, ~{N}-[4-methyl-5-(1-oxidanylidene-7-sulfamoyl-isoindol-5-yl)-1,3-thiazol-2-yl]ethanamide | | Authors: | Petersen, J. | | Deposit date: | 2018-06-27 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A class of highly selective inhibitors bind to an active state of PI3K gamma.
Nat.Chem.Biol., 15, 2019
|
|
1HNV
 
 | | STRUCTURE OF HIV-1 RT(SLASH)TIBO R 86183 COMPLEX REVEALS SIMILARITY IN THE BINDING OF DIVERSE NONNUCLEOSIDE INHIBITORS | | Descriptor: | 5-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Das, K, Ding, J, Arnold, E. | | Deposit date: | 1995-03-30 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of HIV-1 RT/TIBO R 86183 complex reveals similarity in the binding of diverse nonnucleoside inhibitors.
Nat.Struct.Biol., 2, 1995
|
|
