9UY3
 
 | | Anamorelin bound growth hormone secretagogue receptor in complex with Gq | | Descriptor: | 2-azanyl-N-[(2R)-1-[(3S)-3-[dimethylamino(methyl)carbamoyl]-3-(phenylmethyl)piperidin-1-yl]-3-(1H-indol-3-yl)-1-oxidanylidene-propan-2-yl]-2-methyl-propanamide, CHOLESTEROL, Engineered G-alpha-q subunit, ... | | Authors: | Wang, R, Sun, J, Liu, H, Guo, S, Zhang, Y, Hu, W, Wang, J, Liu, H, Zhuang, Y, Jiang, Y, Xie, X, Xu, H.E, Wang, Y. | | Deposit date: | 2025-05-14 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Molecular recognition of two approved drugs Macimorelin and Anamorelin by the growth hormone secretagogue receptor.
Acta Pharmacol.Sin., 2025
|
|
9V2N
 
 | | Macimorelin bound growth hormone secretagogue receptor in complex with Gq | | Descriptor: | Engineered G-alpha-q subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, R, Sun, J, Liu, H, Guo, S, Zhang, Y, Hu, W, Wang, J, Liu, H, Zhuang, Y, Jiang, Y, Xie, X, Xu, H, Wang, Y. | | Deposit date: | 2025-05-20 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | Molecular recognition of two approved drugs Macimorelin and Anamorelin by the growth hormone secretagogue receptor.
Acta Pharmacol.Sin., 2025
|
|
5XP6
 
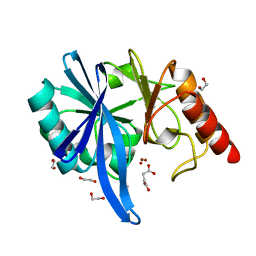 | | native structure of NDM-1 crystallized at pH5.5 | | Descriptor: | 1,2-ETHANEDIOL, HYDROXIDE ION, Metallo-beta-lactamase type 2, ... | | Authors: | Zhang, H, Ma, G, Lai, J, Sun, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Bismuth antimicrobial drugs serve as broad-spectrum metallo-beta-lactamase inhibitors
Nat Commun, 9, 2018
|
|
5XP9
 
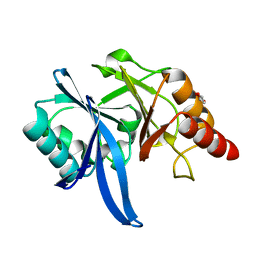 | | Crystal structure of Bismuth bound NDM-1 | | Descriptor: | Bismuth(III) ION, GLYCEROL, Metallo-beta-lactamase type 2 | | Authors: | Zhang, H, Ma, G, Lai, T.P, Sun, H. | | Deposit date: | 2017-06-01 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Bismuth antimicrobial drugs serve as broad-spectrum metallo-beta-lactamase inhibitors
Nat Commun, 9, 2018
|
|
7E8M
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1F11 with mutated RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1F11 heavy chain, ... | | Authors: | Wang, X.Q, Zhang, L.Q, Ge, J.W, Wang, R.K, Lan, J. | | Deposit date: | 2021-03-02 | | Release date: | 2021-05-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species.
Immunity, 54, 2021
|
|
8Y13
 
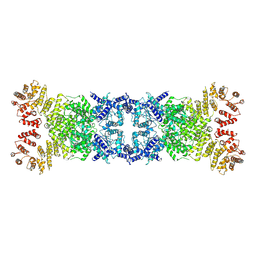 | | Cryo-EM structure of anti-phage defense associated DSR2 tetramer (H171A) | | Descriptor: | SIR2-like domain-containing protein | | Authors: | Li, F.X, Shi, Z.B, Wang, R.W, Xu, Q, Yang, R, Wu, Z.X. | | Deposit date: | 2024-01-23 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y3M
 
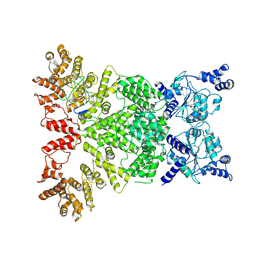 | | Cryo-EM structure of DSR2-DSAD1 complex (cross-linked) | | Descriptor: | DSR anti-defence 1, SIR2-like domain-containing protein | | Authors: | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y3W
 
 | | The Cryo-EM structure of anti-phage defense associated DSR2 tetramer bound with two DSAD1 inhibitors (same side) | | Descriptor: | DSR anti-defence 1, SIR2-like domain-containing protein | | Authors: | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y3Y
 
 | | The Cryo-EM structure of anti-phage defense associated DSR2 tetramer bound with two DSAD1 inhibitors (opposite side) | | Descriptor: | DSR anti-defence 1, SIR2-like domain-containing protein | | Authors: | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
8Y34
 
 | | Cryo-EM structure of anti-phage defense associated DSR2 (H171A) (map2) | | Descriptor: | SIR2-like domain-containing protein | | Authors: | Wang, R.W, Xu, Q, Wu, Z.X, Li, J.L, Shi, Z.B, Li, F.X. | | Deposit date: | 2024-01-28 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | The structural basis of the activation and inhibition of DSR2 NADase by phage proteins.
Nat Commun, 15, 2024
|
|
5GT1
 
 | | crystal structure of cbpa from L. salivarius REN | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Choline binding protein A, ... | | Authors: | Jiang, L, Ren, F. | | Deposit date: | 2016-08-18 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Adhesion of Lactobacillus salivarius REN to a Human Intestinal Epithelial Cell Line Requires S-layer Proteins
Sci Rep, 7, 2017
|
|
5HN2
 
 | |
5HNJ
 
 | |
5HNQ
 
 | |
8XBS
 
 | | C. elegans apo-SID1 structure | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gong, D.S. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Structural basis for double-stranded RNA recognition by SID1.
Nucleic Acids Res., 52, 2024
|
|
8XC1
 
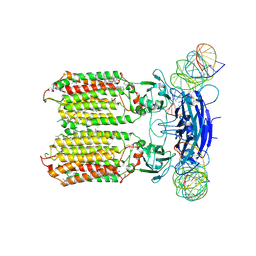 | | C. elegans SID1 in complex with dsRNA | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gong, D.S. | | Deposit date: | 2023-12-07 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Structural basis for double-stranded RNA recognition by SID1.
Nucleic Acids Res., 52, 2024
|
|
6VAF
 
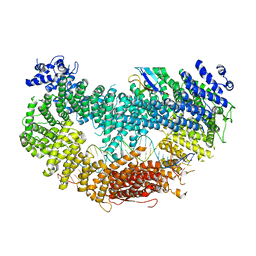 | |
6VAE
 
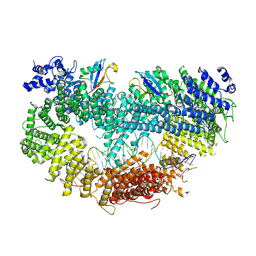 | |
6VAD
 
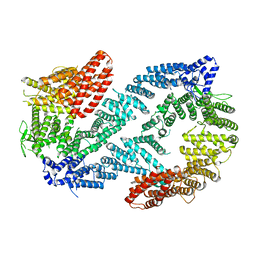 | | Fanconi Anemia ID complex | | Descriptor: | Fanconi anemia group D2 protein, Fanconi anemia, complementation group I | | Authors: | Pavletich, N.P. | | Deposit date: | 2019-12-17 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | DNA clamp function of the monoubiquitinated Fanconi anaemia ID complex.
Nature, 580, 2020
|
|
6VAA
 
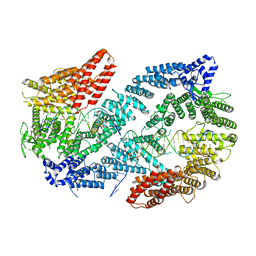 | | Structure of the Fanconi Anemia ID complex bound to ICL DNA | | Descriptor: | DNA (26-MER), DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), ... | | Authors: | Pavletich, N.P. | | Deposit date: | 2019-12-17 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | DNA clamp function of the monoubiquitinated Fanconi anaemia ID complex.
Nature, 580, 2020
|
|
2L8E
 
 | |
8ZCA
 
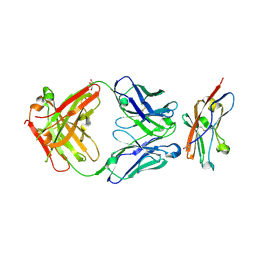 | | Crystal structure of human CD47 ECD bound to Fab of Hu1C8 | | Descriptor: | 1C8 Fab heavy chain, 1C8 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Z, Ding, J. | | Deposit date: | 2024-04-29 | | Release date: | 2025-05-07 | | Last modified: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An anti-CD47 antibody binds to a distinct epitope in a novel metal ion-dependent manner to minimize cross-linking of red blood cells.
J.Biol.Chem., 2025
|
|
8ZYM
 
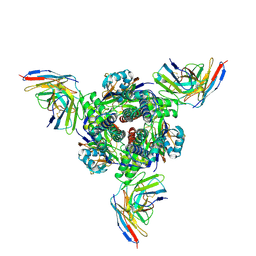 | | Complex structure of 60 Fab bound to DS2 prefusion F trimer | | Descriptor: | Fusion glycoprotein F0,Expression tag, antibody light chain, antidody heavy chain | | Authors: | Wang, X, Ge, J, Guo, F. | | Deposit date: | 2024-06-18 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | DS2 designer pre-fusion F vaccine induces strong and protective antibody response against RSV infection.
Npj Vaccines, 9, 2024
|
|
7MU5
 
 | | Human DCTPP1 bound to Triptolide | | Descriptor: | MAGNESIUM ION, dCTP pyrophosphatase 1, triptolide | | Authors: | Hauk, G, Berger, J.M. | | Deposit date: | 2021-05-14 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Triptolide sensitizes cancer cells to nucleoside DNA methyltransferase inhibitors through inhibition of DCTPP1 mediated cell-intrinsic resistance
To Be Published
|
|
7FJO
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with three T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
