6URO
 
 | | Cryo-EM structure of human CPSF160-WDR33-CPSF30-PAS RNA-CstF77 complex | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 1, Cleavage and polyadenylation specificity factor subunit 4, Cleavage stimulation factor subunit 3, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-10-23 | | Release date: | 2019-11-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Insights into the Human Pre-mRNA 3'-End Processing Machinery.
Mol.Cell, 77, 2020
|
|
4RWW
 
 | | Crystal Structure of L. monocytogenes PstA in complex with cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Lmo2692 protein | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2014-12-07 | | Release date: | 2014-12-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis for the recognition of cyclic-di-AMP by PstA, a PII -like signal transduction protein.
Microbiologyopen, 4, 2015
|
|
6V4X
 
 | | Cryo-EM structure of an active human histone pre-mRNA 3'-end processing machinery at 3.2 Angstrom resolution | | Descriptor: | Cleavage and polyadenylation specificity factor subunit 2, Cleavage and polyadenylation specificity factor subunit 3, Small nuclear ribonucleoprotein E, ... | | Authors: | Sun, Y, Zhang, Y, Walz, T, Tong, L. | | Deposit date: | 2019-12-02 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of an active human histone pre-mRNA 3'-end processing machinery.
Science, 367, 2020
|
|
4RWX
 
 | | Crystal Structure of L. monocytogenes PstA | | Descriptor: | Lmo2692 protein | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2014-12-07 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular basis for the recognition of cyclic-di-AMP by PstA, a PII -like signal transduction protein.
Microbiologyopen, 4, 2015
|
|
4S1B
 
 | | Crystal Structure of L. monocytogenes phosphodiesterase PgpH HD domain in complex with Cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, FE (III) ION, Lmo1466 protein | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2015-01-12 | | Release date: | 2015-02-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6O0H
 
 | |
6O3W
 
 | |
6O3Y
 
 | | Crystal structure of yeast Nrd1 CID in complex with Sen1 NIM3 | | Descriptor: | CHLORIDE ION, Helicase SEN1, Protein NRD1 | | Authors: | Zhang, Y, Tong, L. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Identification of Three Sequence Motifs in the Transcription Termination Factor Sen1 that Mediate Direct Interactions with Nrd1.
Structure, 27, 2019
|
|
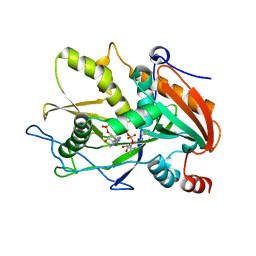
6WUK
 
 | |
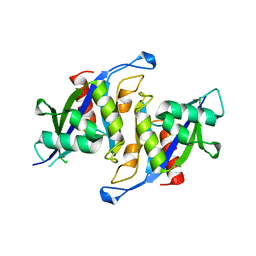
6X7V
 
 | |
6WUG
 
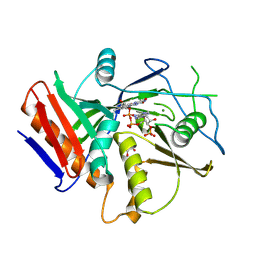 | |
6XGT
 
 | |
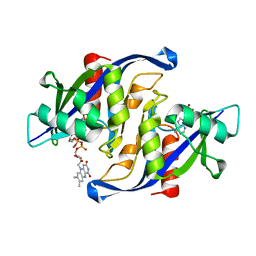
6X7U
 
 | |
6WUI
 
 | | Crystal Structure of mutant S. pombe Rai1 (E150S/E199Q/E239Q) in complex with 3'-FADP | | Descriptor: | Decapping nuclease din1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (2R,3S,4S)-5-(7,8-dimethyl-2,4-dioxo-3,4-dihydrobenzo[g]pteridin-10(2H)-yl)-2,3,4-trihydroxypentyl dihydrogen diphosphate (non-preferred name) | | Authors: | Doamekpor, S.K, Tong, L. | | Deposit date: | 2020-05-04 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DXO/Rai1 enzymes remove 5'-end FAD and dephospho-CoA caps on RNAs.
Nucleic Acids Res., 48, 2020
|
|
6XNV
 
 | | CRYSTAL STRUCTURE OF LISTERIA MONOCYTOGENES CBPB PROTEIN (LMO1009) IN COMPLEX WITH C-DI-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CBS domain-containing protein | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2020-07-04 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | (p)ppGpp and c-di-AMP Homeostasis Is Controlled by CbpB in Listeria monocytogenes.
Mbio, 11, 2020
|
|
6XNU
 
 | |
5BTE
 
 | |
5BTH
 
 | | Crystal structure of Candida albicans Rai1 | | Descriptor: | Decapping nuclease RAI1 | | Authors: | Wang, V.Y, Tong, L. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical studies of the distinct activity profiles of Rai1 enzymes.
Nucleic Acids Res., 43, 2015
|
|
5BUD
 
 | |
5BTO
 
 | |
5BTB
 
 | | Crystal Structure of Ashbya gossypii Rai1 | | Descriptor: | AFR263Cp, SULFATE ION | | Authors: | Wang, V.Y, Tong, L. | | Deposit date: | 2015-06-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and biochemical studies of the distinct activity profiles of Rai1 enzymes.
Nucleic Acids Res., 43, 2015
|
|
8UIB
 
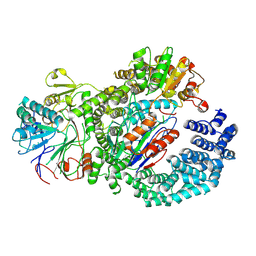 | | Structure of the human INTS9-INTS11-BRAT1 complex | | Descriptor: | BRCA1-associated ATM activator 1, Integrator complex subunit 11, Integrator complex subunit 9, ... | | Authors: | Lin, M, Tong, L. | | Deposit date: | 2023-10-10 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cytoplasmic binding partners of the Integrator endonuclease INTS11 and its paralog CPSF73 are required for their nuclear function.
Mol.Cell, 84, 2024
|
|
8UIC
 
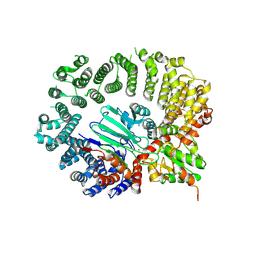 | | Structure of the Drosophila IntS11-CG7044(dBRAT1) complex | | Descriptor: | FI02071p, Integrator complex subunit 11, ZINC ION | | Authors: | Lin, M, Tong, L. | | Deposit date: | 2023-10-10 | | Release date: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Cytoplasmic binding partners of the Integrator endonuclease INTS11 and its paralog CPSF73 are required for their nuclear function.
Mol.Cell, 84, 2024
|
|
4FGM
 
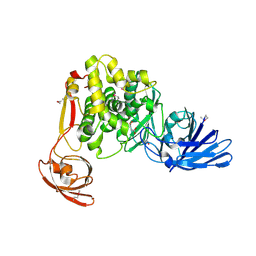 | | Crystal structure of the aminopeptidase N family protein Q5QTY1 from Idiomarina loihiensis. Northeast Structural Genomics Consortium Target IlR60. | | Descriptor: | Aminopeptidase N family protein, MALEIC ACID, ZINC ION | | Authors: | Vorobiev, S, Su, M, Tong, T, Kohan, E, Wang, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-04 | | Release date: | 2012-08-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Crystal structure of the aminopeptidase N family protein Q5QTY1 from Idiomarina loihiensis.
To be Published
|
|
3B57
 
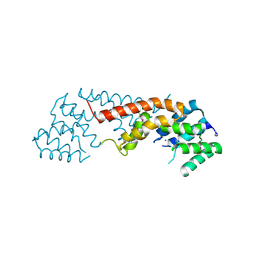 | | Crystal structure of the Lin1889 protein (Q92AN1) from Listeria innocua. Northeast Structural Consortium target LkR65 | | Descriptor: | Lin1889 protein, MAGNESIUM ION | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Benach, J, Forouhar, F, Vorobiev, S.M, Wang, H, Janjua, H, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-25 | | Release date: | 2007-11-06 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Lin1889 protein (Q92AN1) from Listeria innocua.
To be Published
|
|
