7BE1
 
 | | X-ray structure of Hen Egg White Lysozyme with dirhodium tetraacetate (3) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Lysozyme, ... | | Authors: | Loreto, D, Merlino, A, Ferraro, G. | | Deposit date: | 2020-12-22 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Unusual Structural Features in the Adduct of Dirhodium Tetraacetate with Lysozyme.
Int J Mol Sci, 22, 2021
|
|
7BE0
 
 | | X-ray structure of Hen Egg White Lysozyme with dirhodium tetraacetate (2) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Lysozyme, ... | | Authors: | Loreto, D, Merlino, A, Ferraro, G. | | Deposit date: | 2020-12-22 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Unusual Structural Features in the Adduct of Dirhodium Tetraacetate with Lysozyme.
Int J Mol Sci, 22, 2021
|
|
3M7Q
 
 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Caribbean sea anemone stichodactyla helianthus in complex with bovine pancreatic trypsin | | Descriptor: | Cationic trypsin, Kunitz-type proteinase inhibitor SHPI-1, PHOSPHATE ION | | Authors: | Garcia-Fernandez, R, Redecke, L, Pons, T, Perbandt, M, Gil, D, Talavera, A, Gonzalez, Y, de los angeles Chavez, M, Betzel, C. | | Deposit date: | 2010-03-17 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insights into serine protease inhibition by a marine invertebrate BPTI Kunitz-type inhibitor.
J.Struct.Biol., 180, 2012
|
|
8RPM
 
 | |
8QB1
 
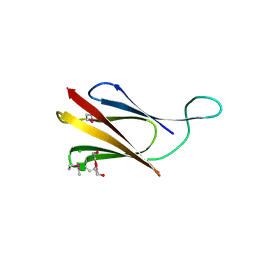 | | C-terminal domain of mirolase from Tannerella forsythia | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Mirolase, ... | | Authors: | Gomis-Ruth, F.X, Rodriguez-Banqueri, A, Mizgalska, D, Veillard, F, Goulas, T, Eckhard, U, Potempa, J. | | Deposit date: | 2023-08-23 | | Release date: | 2024-02-28 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and functional insights into the C-terminal signal domain of the Bacteroidetes type-IX secretion system.
Open Biology, 14, 2024
|
|
7BC3
 
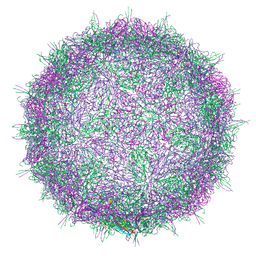 | | Native virion of Kashmir bee virus at acidic pH | | Descriptor: | Structural polyprotein | | Authors: | Mukhamedova, L, Plevka, P, Fuzik, T, Hrebik, D, Novacek, J. | | Deposit date: | 2020-12-18 | | Release date: | 2021-03-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Virion structure and in vitro genome release mechanism of dicistrovirus Kashmir bee virus.
J.Virol., 95, 2021
|
|
6ZQJ
 
 | | Cryo-EM structure of trimeric prME spike of Spondweni virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein, prM | | Authors: | Renner, M, Dejnirattisai, W, Carrique, L, Serna Martin, I, Karia, D, Ilca, S.L, Ho, S.F, Kotecha, A, Keown, J.R, Mongkolsapaya, J, Screaton, G.R, Grimes, J.M. | | Deposit date: | 2020-07-09 | | Release date: | 2021-01-20 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins.
Nat Commun, 12, 2021
|
|
6ZQV
 
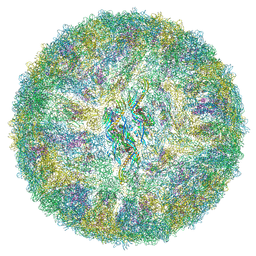 | | Cryo-EM structure of mature Spondweni virus | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Genome polyprotein | | Authors: | Renner, M, Dejnirattisai, W, Carrique, L, Serna Martin, I, Karia, D, Ilca, S.L, Ho, S.F, Kotecha, A, Keown, J.R, Mongkolsapaya, J, Screaton, G.R, Grimes, J.M. | | Deposit date: | 2020-07-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Flavivirus maturation leads to the formation of an occupied lipid pocket in the surface glycoproteins.
Nat Commun, 12, 2021
|
|
6ZWO
 
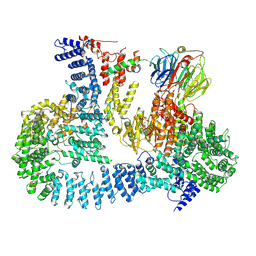 | | cryo-EM structure of human mTOR complex 2, focused on one half | | Descriptor: | ACETYL GROUP, INOSITOL HEXAKISPHOSPHATE, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Scaiola, A, Mangia, F, Imseng, S, Boehringer, D, Ban, N, Maier, T. | | Deposit date: | 2020-07-28 | | Release date: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The 3.2- angstrom resolution structure of human mTORC2.
Sci Adv, 6, 2020
|
|
8R4I
 
 | |
7A2D
 
 | | Structure-function analyses of dual-BON domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation to the cell division site | | Descriptor: | Uncharacterized protein YraP | | Authors: | Bryant, J.A, Morris, F.C, Knowles, T.J, Maderbocus, R, Heinz, E, Boelter, G, Alodaini, D, Colyer, A, Wotherspoon, P.J, Staunton, K.A, Jeeves, M, Browning, D.F, Sevastsyanovich, Y.R, Wells, T.J, Rossiter, A.E, Bavro, V.N, Sridhar, P, Ward, D.G, Chong, Z.S, Goodall, E.C.A, Icke, C, Teo, A, Chng, S.S, Roper, D.I, Lithgow, T, Cunningham, A.F, Banzhaf, M, Overduin, M, Henderson, I.R. | | Deposit date: | 2020-08-17 | | Release date: | 2020-12-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of dual BON-domain protein DolP identifies phospholipid binding as a new mechanism for protein localisation.
Elife, 9, 2020
|
|
7A77
 
 | | Crystal structure of RXR alpha LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Nuclear receptor coactivator 2, ... | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comprehensive Set of Tertiary Complex Structures and Palmitic Acid Binding Provide Molecular Insights into Ligand Design for RXR Isoforms.
Int J Mol Sci, 21, 2020
|
|
7A79
 
 | | Crystal structure of RXR gamma LBD in complexes with palmitic acid and GRIP-1 peptide | | Descriptor: | Nuclear receptor coactivator 2, PALMITIC ACID, Retinoic acid receptor RXR-gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Comprehensive Set of Tertiary Complex Structures and Palmitic Acid Binding Provide Molecular Insights into Ligand Design for RXR Isoforms.
Int J Mol Sci, 21, 2020
|
|
7AAB
 
 | | Crystal structure of the catalytic domain of human PARP1 in complex with inhibitor EB-47 | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Poly [ADP-ribose] polymerase 1, SULFATE ION | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Easton, L.E, Underwood, E, Rawlins, P.B, Johannes, J.W, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
8C0E
 
 | | The lipid linked oligosaccharide polymerase Wzy and its regulating co-polymerase Wzz form a complex in vivo and in vitro | | Descriptor: | ECA polysaccharide chain length modulation protein | | Authors: | Weckener, M, Woodward, L.S, Clarke, B.R, Liu, H, Ward, P.N, Le Bas, A, Bhella, D, Whitfield, C, Naismith, J.H. | | Deposit date: | 2022-12-16 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The lipid linked oligosaccharide polymerase Wzy and its regulating co-polymerase, Wzz, from enterobacterial common antigen biosynthesis form a complex.
Open Biology, 13, 2023
|
|
6ZZ1
 
 | | Crystal structure of MLKL executioner domain in complex with a covalent inhibitor | | Descriptor: | 7-(2-methoxyethoxymethyl)-1,3-dimethyl-purine-2,6-dione, Mixed lineage kinase domain-like protein | | Authors: | Fiegen, D, Bauer, M, Nar, H. | | Deposit date: | 2020-08-03 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Locking mixed-lineage kinase domain-like protein in its auto-inhibited state prevents necroptosis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8PVA
 
 | | Structure of bacterial ribosome determined by cryoEM at 100 keV | | Descriptor: | 16S rRNA, 23S rRNA, 50S ribosomal protein L14, ... | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVG
 
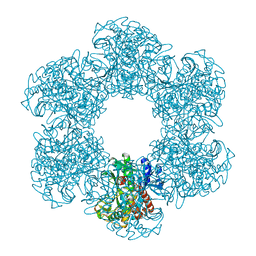 | | Structure of E. coli glutamine synthetase determined by cryoEM at 100 keV | | Descriptor: | Glutamine synthetase | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7A4M
 
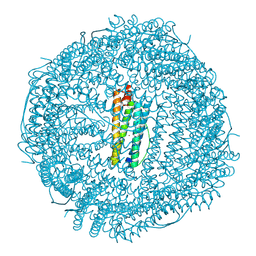 | | Cryo-EM structure of mouse heavy-chain apoferritin at 1.22 A | | Descriptor: | FE (III) ION, Ferritin heavy chain, ZINC ION | | Authors: | Nakane, T, Kotecha, A, Sente, A, Yamashita, K, McMullan, G, Masiulis, S, Brown, P.M.G.E, Grigoras, I.T, Malinauskaite, L, Malinauskas, T, Miehling, J, Yu, L, Karia, D, Pechnikova, E.V, de Jong, E, Keizer, J, Bischoff, M, McCormack, J, Tiemeijer, P, Hardwick, S.W, Chirgadze, D.Y, Murshudov, G, Aricescu, A.R, Scheres, S.H.W. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (1.22 Å) | | Cite: | Single-particle cryo-EM at atomic resolution.
Nature, 587, 2020
|
|
7A5V
 
 | | CryoEM structure of a human gamma-aminobutyric acid receptor, the GABA(A)R-beta3 homopentamer, in complex with histamine and megabody Mb25 in lipid nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Nakane, T, Kotecha, A, Sente, A, Yamashita, K, McMullan, G, Masiulis, S, Brown, P.M.G.E, Grigoras, I.T, Malinauskaite, L, Malinauskas, T, Miehling, J, Yu, L, Karia, D, Pechnikova, E.V, de Jong, E, Keizer, J, Bischoff, M, McCormack, J, Tiemeijer, P, Hardwick, S.W, Chirgadze, D.Y, Murshudov, G, Aricescu, A.R, Scheres, S.H.W. | | Deposit date: | 2020-08-22 | | Release date: | 2020-11-18 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (1.7 Å) | | Cite: | Single-particle cryo-EM at atomic resolution.
Nature, 587, 2020
|
|
8PM6
 
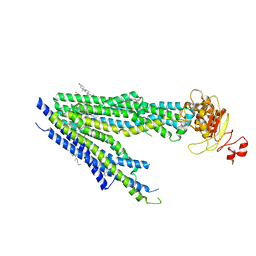 | | Human bile salt export pump (BSEP) in complex with inhibitor GBM in nanodiscs | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, Bile salt export pump, CHOLESTEROL | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
7AAA
 
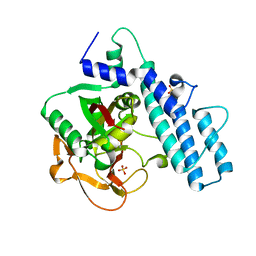 | | Crystal structure of the catalytic domain of human PARP1 (apo) | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Poly [ADP-ribose] polymerase 1, ... | | Authors: | Schimpl, M, Ogden, T.E.H, Yang, J.-C, Underwood, E, Rawlins, P.B, Johannes, J.W, Easton, L.E, Embrey, K.J, Neuhaus, D. | | Deposit date: | 2020-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Dynamics of the HD regulatory subdomain of PARP-1; substrate access and allostery in PARP activation and inhibition.
Nucleic Acids Res., 49, 2021
|
|
6ZML
 
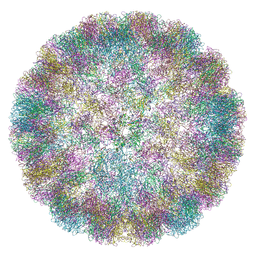 | | CryoEM Structure of Merkel Cell Polyomavirus Virus-like Particle | | Descriptor: | Capsid protein VP1 | | Authors: | Bayer, N.J, Januliene, D, Stehle, T, Moeller, A, Blaum, B.S. | | Deposit date: | 2020-07-03 | | Release date: | 2020-08-05 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of Merkel Cell Polyomavirus Capsid and Interaction with Its Glycosaminoglycan Attachment Receptor.
J.Virol., 94, 2020
|
|
8C0C
 
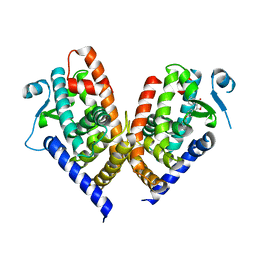 | | X-ray crystal structure of PPAR gamma ligand binding domain in complex with CZ46 | | Descriptor: | (2~{R})-2-[4-(naphthalen-1-ylmethoxy)phenyl]-4-oxidanyl-3-phenyl-2~{H}-furan-5-one, Peroxisome proliferator-activated receptor gamma | | Authors: | Capelli, D, Montanari, R, Pochetti, G, Villa, S, Meneghetti, F. | | Deposit date: | 2022-12-16 | | Release date: | 2023-04-26 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biological Screening and Crystallographic Studies of Hydroxy gamma-Lactone Derivatives to Investigate PPAR gamma Phosphorylation Inhibition.
Biomolecules, 13, 2023
|
|
3MS6
 
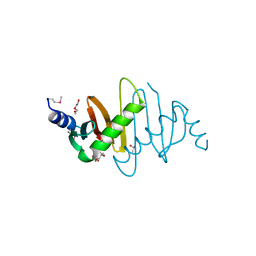 | | Crystal structure of Hepatitis B X-Interacting Protein (HBXIP) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Hepatitis B virus X-interacting protein, ... | | Authors: | Garcia-Saez, I, Skoufias, D. | | Deposit date: | 2010-04-29 | | Release date: | 2010-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Structural Characterization of HBXIP: The Protein That Interacts with the Anti-Apoptotic Protein Survivin and the Oncogenic Viral Protein HBx.
J.Mol.Biol., 405, 2011
|
|
