3BXV
 
 | |
6KMH
 
 | | The crystal structure of CASK/Mint1 complex | | Descriptor: | Amyloid-beta A4 precursor protein-binding family A member 1, CHLORIDE ION, IODIDE ION, ... | | Authors: | Li, W, Feng, W. | | Deposit date: | 2019-07-31 | | Release date: | 2020-08-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CASK modulates the assembly and function of the Mint1/Munc18-1 complex to regulate insulin secretion.
Cell Discov, 6, 2020
|
|
4HYP
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | DNA gyrase subunit B, MAGNESIUM ION, N-[7-(1H-imidazol-1-yl)-2-(pyridin-3-yl)[1,3]thiazolo[5,4-d]pyrimidin-5-yl]cyclopropanecarboxamide | | Authors: | Bensen, D.C, Creighton, C.J, Tari, L.W. | | Deposit date: | 2012-11-13 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HZ5
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity | | Descriptor: | 6-ethyl-4-methoxy-2-(pyridin-3-ylsulfanyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbaldehyde, DNA topoisomerase IV, B subunit | | Authors: | Bensen, D.C, Creighton, C.J, Tari, L.W. | | Deposit date: | 2012-11-14 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6KK0
 
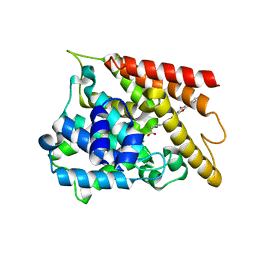 | | Crystal structure of PDE4D catalytic domain complexed with compound 4e | | Descriptor: | 7-[8-methoxy-2,2-dimethyl-7-(3-methylbut-2-enyl)-5-oxidanyl-6-oxidanylidene-pyrano[3,2-b]xanthen-9-yl]oxyheptanoic acid, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2019-07-23 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.20008755 Å) | | Cite: | Discovery and Optimization of alpha-Mangostin Derivatives as Novel PDE4 Inhibitors for the Treatment of Vascular Dementia.
J.Med.Chem., 63, 2020
|
|
2E9N
 
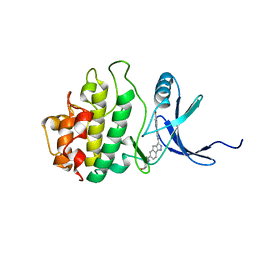 | | Structure of h-CHK1 complexed with A767085 | | Descriptor: | 3-(4'-HYDROXYBIPHENYL-4-YL)-N-(4-HYDROXYCYCLOHEXYL)-1,4-DIHYDROINDENO[1,2-C]PYRAZOLE-6-CARBOXAMIDE, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-01-26 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of 1,4-dihydroindeno[1,2-c]pyrazoles as a novel class of potent and selective checkpoint kinase 1 inhibitors.
Bioorg.Med.Chem., 15, 2007
|
|
3SKO
 
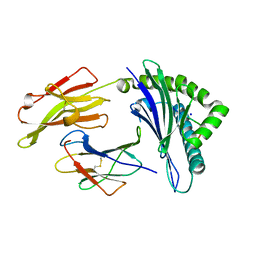 | | Crystal structure of the HLA-B8-A66-FLR, mutant A66 of the HLA B8 | | Descriptor: | Beta-2-microglobulin, Epstein-Barr nuclear antigen 3, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Wilmann, P.G, Zhenjun, C, Hanim, H, Yu Chih, L, Kjer-Nielsen, L, Purcell, A.W, Burrows, S.R, Mccluskey, J, Rossjohn, J. | | Deposit date: | 2011-06-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A structural basis for varied alpha-beta TCR usage against an immunodominant EBV antigen restricted to a HLA-B8 molecule.
J.Immunol., 188, 2012
|
|
3SKN
 
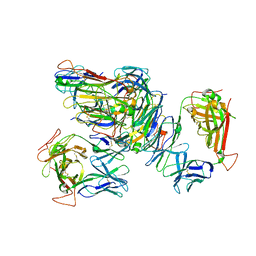 | | Crystal structure of the RL42 TCR unliganded | | Descriptor: | RL42 T cell receptor, alpha chain, beta chain | | Authors: | Gras, S, Wilmann, P.G, Zhenjun, C, Hanim, H, Yu Chih, L, Kjer-Nielsen, L, Purcell, A.W, Burrows, S.R, Mccluskey, J, Rossjohn, J. | | Deposit date: | 2011-06-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A structural basis for varied alpha-beta TCR usage against an immunodominant EBV antigen restricted to a HLA-B8 molecule.
J.Immunol., 188, 2012
|
|
6KW3
 
 | | The ClassA RSC-Nucleosome Complex | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-09-05 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.13 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
2E9O
 
 | | Structure of h-CHK1 complexed with AA582939 | | Descriptor: | 4-(6-{[(4-METHYLCYCLOHEXYL)AMINO]METHYL}-1,4-DIHYDROINDENO[1,2-C]PYRAZOL-3-YL)BENZOIC ACID, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-01-26 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of 1,4-dihydroindeno[1,2-c]pyrazoles as a novel class of potent and selective checkpoint kinase 1 inhibitors.
Bioorg.Med.Chem., 15, 2007
|
|
4Y19
 
 | | immune complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FS18_alpha, FS18_beta, ... | | Authors: | Beringer, D.X, Petersen, J, Reid, H.H, Rossjohn, J. | | Deposit date: | 2015-02-07 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex.
Nat.Immunol., 16, 2015
|
|
6L30
 
 | | Crystal structure of the epithelial cell transforming 2 (ECT2) | | Descriptor: | Protein ECT2 | | Authors: | Chen, Z.C, Chen, M.R, Pan, H, Sun, L.F, Shi, P. | | Deposit date: | 2019-10-07 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and regulation of human epithelial cell transforming 2 protein.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2E9V
 
 | | Structure of h-CHK1 complexed with A859017 | | Descriptor: | 18-CHLORO-2-OXO-17-[(PYRIDIN-4-YLMETHYL)AMINO]-2,3,11,12,13,14-HEXAHYDRO-1H,10H-4,8-(AZENO)-9,15,1,3,6-BENZODIOXATRIAZACYCLOHEPTADECINE-7-CARBONITRILE, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-01-27 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design, Synthesis and Biological Evaluation of Potent Selective Macrocyclic Chk1 Inhibitors
To be Published
|
|
2E9P
 
 | | Structure of h-CHK1 complexed with A771129 | | Descriptor: | 1-(5-CHLORO-2-METHOXYPHENYL)-3-{6-[2-(DIMETHYLAMINO)-1-METHYLETHOXY]PYRAZIN-2-YL}UREA, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-01-26 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design, Synthesis and Biological Evaluation of Potent and Selective Macrocyclic Chk1 Inhibitors
To be Published
|
|
2E9U
 
 | | Structure of h-CHK1 complexed with A780125 | | Descriptor: | 18-CHLORO-11,12,13,14-TETRAHYDRO-1H,10H-8,4-(AZENO)-9,15,1,3,6-BENZODIOXATRIAZACYCLOHEPTADECIN-2-ONE, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-01-27 | | Release date: | 2008-01-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design, Synthesis and Biological Evaluation of Potent Selective Macrocyclic Chk1 Inhibitors
To be Published
|
|
3SRU
 
 | | S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines | | Descriptor: | Dihydrofolate reductase, N-[3'-(2,4-diaminoquinazolin-7-yl)-4'-ethoxybiphenyl-3-yl]methanesulfonamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M. | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
7WX6
 
 | | A Legionella acetyltransferase VipF | | Descriptor: | CHLORAMPHENICOL, COENZYME A, N-acetyltransferase | | Authors: | Chen, T.T, Lin, Y.L, Chen, Z, Han, A.D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.273 Å) | | Cite: | Structural basis for the acetylation mechanism of the Legionella effector VipF.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7ELG
 
 | | LC3B modificated with a covalent probe | | Descriptor: | 2-methylidene-5-thiophen-2-yl-cyclohexane-1,3-dione, Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | Authors: | Fan, S, Wan, W. | | Deposit date: | 2021-04-10 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Inhibition of Autophagy by a Small Molecule through Covalent Modification of the LC3 Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4Y1A
 
 | | immune complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FS17_alpha, FS17_beta, ... | | Authors: | Beringer, D.X, Vivian, J.P, Reid, H.H, Rossjohn, J. | | Deposit date: | 2015-02-07 | | Release date: | 2015-09-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | T cell receptor reversed polarity recognition of a self-antigen major histocompatibility complex.
Nat.Immunol., 16, 2015
|
|
6L6L
 
 | | Structural basis of NR4A2 homodimers binding to selective Nur-responsive elements | | Descriptor: | DNA (5'-D(*AP*AP*AP*GP*GP*TP*CP*AP*AP*AP*CP*TP*GP*TP*GP*AP*CP*CP*TP*AP*T)-3'), DNA (5'-D(P*TP*AP*TP*AP*GP*GP*TP*CP*AP*CP*AP*GP*TP*TP*TP*GP*AP*CP*CP*TP*T)-3'), Nuclear receptor related 1, ... | | Authors: | Jiang, L, Chen, Y. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Structural basis of binding of homodimers of the nuclear receptor NR4A2 to selective Nur-responsive DNA elements.
J.Biol.Chem., 294, 2019
|
|
3SRS
 
 | | S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines | | Descriptor: | 7-(5-bromo-2-ethoxyphenyl)-6-methylquinazoline-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M. | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6L8L
 
 | | C-Src in complex with ibrutinib | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Guo, M, Dai, S, Chen, L, Chen, Y. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.888 Å) | | Cite: | Characterization of ibrutinib as a non-covalent inhibitor of SRC-family kinases.
Bioorg.Med.Chem.Lett., 34, 2020
|
|
4JQX
 
 | | HLA-B*44:03 in complex with Epstein-Barr virus BZLF1-derived peptide (residues 169-180) | | Descriptor: | ACETATE ION, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Theodossis, A, Welland, A, Gras, S, Rossjohn, J. | | Deposit date: | 2013-03-20 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HLA Peptide Length Preferences Control CD8+ T Cell Responses.
J.Immunol., 191, 2013
|
|
3SRQ
 
 | | S. aureus Dihydrofolate Reductase complexed with novel 7-aryl-2,4-diaminoquinazolines | | Descriptor: | 1-[3-(2,4-diamino-6-methylquinazolin-7-yl)phenyl]ethanone, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M. | | Deposit date: | 2011-07-07 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-based design of new DHFR-based antibacterial agents: 7-aryl-2,4-diaminoquinazolines.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
6L6Q
 
 | |
