6C79
 
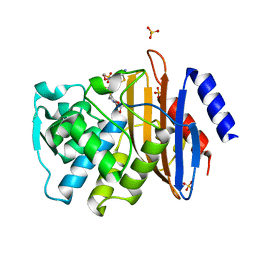 | | Conformational Changes in a Class A Beta lactamase that Prime it for Catalysis | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, Beta-lactamase Toho-1, SULFATE ION | | Authors: | Coates, L, Langan, P.S, Vandavasi, V.G, Cooper, S.J, Weiss, K.L, Ginell, S.L, Parks, J.M. | | Deposit date: | 2018-01-22 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Substrate Binding Induces Conformational Changes in a Class A Beta-lactamase That Prime It for Catalysis
Acs Catalysis, 8, 2018
|
|
1XQ4
 
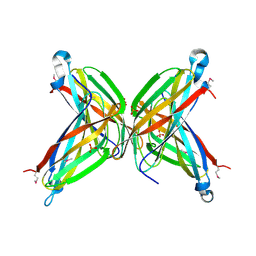 | | Crystal Structure of the Putative ApaA Protein from Bordetella pertussis, Northeast Structural Genomics Target BeR40 | | Descriptor: | PHOSPHATE ION, Protein apaG | | Authors: | Forouhar, F, Yong, W, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-11 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Putative ApaA Protein from Bordetella pertussis, Northeast Structural Genomics Target BeR40
To be Published
|
|
1RSW
 
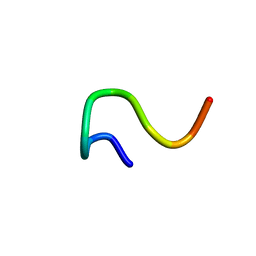 | | 12-mer from site II calbindin D9K (DKNGDGEVSFEE) coordination Pb(II) | | Descriptor: | Vitamin D-dependent calcium-binding protein, intestinal | | Authors: | Ferrari, R, Mendoza, G, James, T, Tonelli, M, Basus, V, Gasque, L. | | Deposit date: | 2003-12-10 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Coordination Compounds derived from a dodecapeptide and Pb2+, Cd2+ and Zn2+. Modeling the site II from the Calbindin D9K
To be published
|
|
1XQE
 
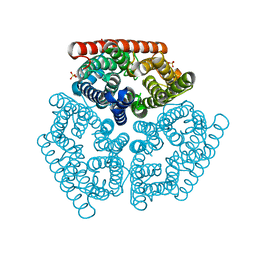 | | The mechanism of ammonia transport based on the crystal structure of AmtB of E. coli. | | Descriptor: | ACETATE ION, Probable ammonium transporter, SULFATE ION | | Authors: | Zheng, L, Kostrewa, D, Berneche, S, Winkler, F.K, Li, X.-D. | | Deposit date: | 2004-10-12 | | Release date: | 2004-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6C92
 
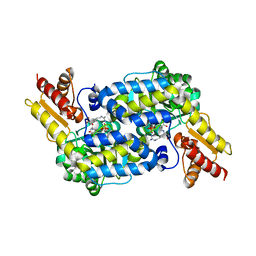 | | The structure of MppP soaked with the product 2-ketoarginine | | Descriptor: | (4S)-5-carbamimidamido-4-hydroxy-2-oxopentanoic acid, (E)-N~2~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-arginine, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-25 | | Release date: | 2018-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.834 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
8PMD
 
 | | Nucleotide-bound BSEP in nanodiscs | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bile salt export pump, MAGNESIUM ION | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
1XR1
 
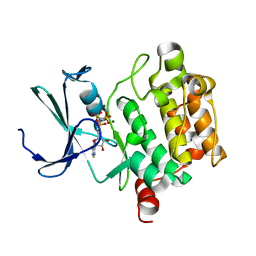 | | Crystal structure of hPim-1 kinase in complex with AMP-PNP at 2.1 A Resolution | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Qian, K.C, Wang, L, Hickey, E.R, Studts, J, Barringer, K, Peng, C, Kronkaitis, A, Li, J, White, A, Mische, S, Farmer, B. | | Deposit date: | 2004-10-13 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Constitutive Activity and a Unique Nucleotide Binding Mode of Human Pim-1 Kinase.
J.Biol.Chem., 280, 2005
|
|
6C9B
 
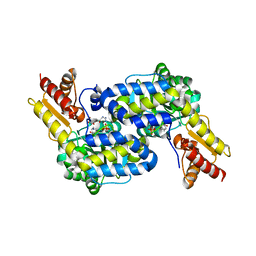 | | The structure of MppP soaked with the products 4HKA and 2KA | | Descriptor: | (4S)-5-carbamimidamido-4-hydroxy-2-oxopentanoic acid, CHLORIDE ION, PLP-Dependent L-Arginine Hydroxylase MppP | | Authors: | Han, L, Silvaggi, N.R. | | Deposit date: | 2018-01-26 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.689 Å) | | Cite: | Streptomyces wadayamensis MppP is a PLP-Dependent Oxidase, Not an Oxygenase.
Biochemistry, 57, 2018
|
|
8PMJ
 
 | | Vanadate-trapped BSEP in nanodiscs | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Bile salt export pump, ... | | Authors: | Liu, H, Irobalieva, R.N, Kowal, J, Ni, D, Nosol, K, Bang-Sorensen, R, Lancien, L, Stahlberg, H, Stieger, B, Locher, K.P. | | Deposit date: | 2023-06-28 | | Release date: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural basis of bile salt extrusion and small-molecule inhibition in human BSEP.
Nat Commun, 14, 2023
|
|
2NZ2
 
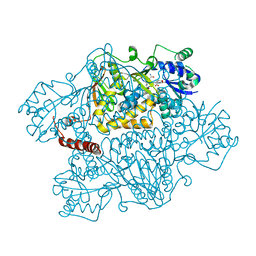 | | Crystal structure of human argininosuccinate synthase in complex with aspartate and citrulline | | Descriptor: | ASPARTIC ACID, Argininosuccinate synthase, CITRULLINE, ... | | Authors: | Karlberg, T, Uppenberg, J, Arrowsmith, C, Berglund, H, Busam, R.D, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Hogbom, M, Johansson, I, Kotenyova, T, Magnusdottir, A, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-22 | | Release date: | 2006-12-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human argininosuccinate synthetase.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6CAE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with NOSO-95179 antibiotic and bound to mRNA and A-, P- and E-site tRNAs at 2.6A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Pantel, L, Florin, T, Dobosz-Bartoszek, M, Racine, E, Sarciaux, M, Serri, M, Houard, J, Campagne, J.M, Marcia de Figueiredo, R, Midrier, C, Gaudriault, S, Givaudan, A, Lanois, A, Forst, S, Aumelas, A, Cotteaux-Lautard, C, Bolla, J.M, Vingsbo Lundberg, C, Huseby, D, Hughes, D, Villain-Guillot, P, Mankin, A.S, Polikanov, Y.S, Gualtieri, M. | | Deposit date: | 2018-01-30 | | Release date: | 2018-04-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Odilorhabdins, Antibacterial Agents that Cause Miscoding by Binding at a New Ribosomal Site.
Mol. Cell, 70, 2018
|
|
8PN8
 
 | | Engineered glycolyl-CoA carboxylase (L100N variant) with bound CoA | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Propionyl-CoA carboxylase alpha subunit, ... | | Authors: | Zarzycki, J, Marchal, D.G, Schulz, L, Prinz, S, Erb, T.J. | | Deposit date: | 2023-06-30 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.31 Å) | | Cite: | Machine Learning-Supported Enzyme Engineering toward Improved CO 2 -Fixation of Glycolyl-CoA Carboxylase.
Acs Synth Biol, 12, 2023
|
|
1RW2
 
 | | Three-dimensional structure of Ku80 CTD | | Descriptor: | ATP-dependent DNA helicase II, 80 kDa subunit | | Authors: | Zhang, Z, Hu, W, Cano, L, Lee, T.D, Chen, D.J, Chen, Y. | | Deposit date: | 2003-12-15 | | Release date: | 2003-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions.
STRUCTURE, 12, 2004
|
|
1XTO
 
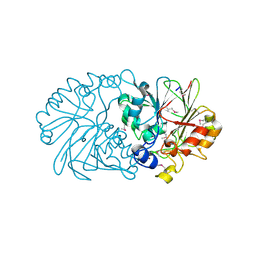 | | Crystal Structure of the Coenzyme PQQ Synthesis Protein (PqqB) from Pseudomonas putida, Northeast Structural Genomics Target PpR6 | | Descriptor: | Coenzyme PQQ synthesis protein B, ZINC ION | | Authors: | Forouhar, F, Chen, Y, Kuzin, A, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-22 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Coenzyme PQQ Synthesis Protein (PqqB) from Pseudomonas putida, Northeast Structural Genomics Target PpR6
To be Published
|
|
8PDP
 
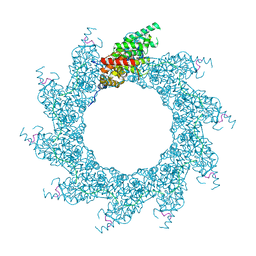 | | 10-mer ring of HMPV N-RNA bound to the C-terminal region of P | | Descriptor: | Nucleoprotein, Phosphoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
1REO
 
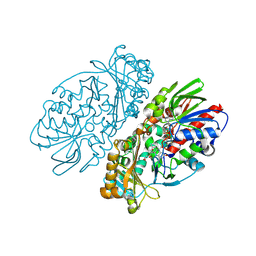 | | L-amino acid oxidase from Agkistrodon halys pallas | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AHPLAAO, CITRIC ACID, ... | | Authors: | Zhang, H, Teng, M, Niu, L, Wang, Y, Wang, Y, Liu, Q, Huang, Q, Hao, Q, Dong, Y, Liu, P. | | Deposit date: | 2003-11-07 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Purification, partial characterization, crystallization and structural determination of AHP-LAAO, a novel L-amino-acid oxidase with cell apoptosis-inducing activity from Agkistrodon halys pallas venom.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6C5W
 
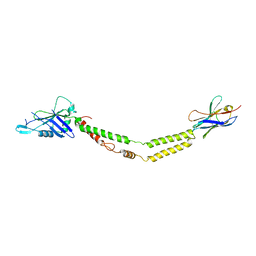 | | Crystal structure of the mitochondrial calcium uniporter | | Descriptor: | CALCIUM ION, calcium uniporter, nanobody | | Authors: | Fan, C, Fan, M, Fastman, N, Zhang, J, Feng, L. | | Deposit date: | 2018-01-17 | | Release date: | 2018-07-11 | | Last modified: | 2019-04-24 | | Method: | X-RAY DIFFRACTION (3.10010242 Å) | | Cite: | X-ray and cryo-EM structures of the mitochondrial calcium uniporter.
Nature, 559, 2018
|
|
1R4A
 
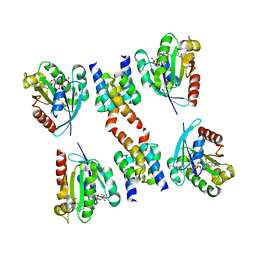 | | Crystal Structure of GTP-bound ADP-ribosylation Factor Like Protein 1 (Arl1) and GRIP Domain of Golgin245 COMPLEX | | Descriptor: | ADP-ribosylation factor-like protein 1, Golgi autoantigen, golgin subfamily A member 4, ... | | Authors: | Wu, M, Lu, L, Hong, W, Song, H. | | Deposit date: | 2003-10-04 | | Release date: | 2004-01-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for recruitment of GRIP domain golgin-245 by small GTPase Arl1.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1X8E
 
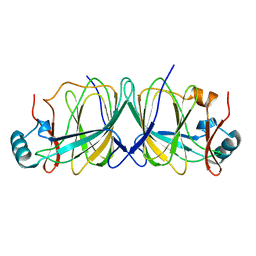 | | Crystal structure of Pyrococcus furiosus phosphoglucose isomerase free enzyme | | Descriptor: | Glucose-6-phosphate isomerase | | Authors: | Berrisford, J.M, Akerboom, J, Brouns, S, Sedelnikova, S.E, Turnbull, A.P, van der Oost, J, Salmon, L, Hardre, R, Murray, I.A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structures of inhibitor complexes of Pyrococcus furiosus phosphoglucose isomerase provide insights into substrate binding and catalysis.
J.Mol.Biol., 343, 2004
|
|
8PDM
 
 | | 11-mer ring of human metapneumovirus (HMPV) N-RNA | | Descriptor: | Nucleoprotein, RNA | | Authors: | Whitehead, J.D, Decool, H, Leyrat, C, Carrique, L, Fix, J, Eleouet, J.F, Galloux, M, Renner, M. | | Deposit date: | 2023-06-12 | | Release date: | 2023-12-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the N-RNA/P interface indicates mode of L/P recruitment to the nucleocapsid of human metapneumovirus.
Nat Commun, 14, 2023
|
|
5DJ5
 
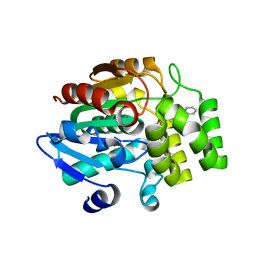 | | Crystal structure of rice DWARF14 in complex with synthetic strigolactone GR24 | | Descriptor: | (3E,3aR,8bS)-3-({[(2R)-4-methyl-5-oxo-2,5-dihydrofuran-2-yl]oxy}methylidene)-3,3a,4,8b-tetrahydro-2H-indeno[1,2-b]furan-2-one, Probable strigolactone esterase D14 | | Authors: | Zhou, X.E, Zhao, L.-H, Yi, W, Wu, Z.-S, Liu, Y, Kang, Y, Hou, L, de Waal, P.W, Li, S, Jiang, Y, Melcher, K, Xu, H.E. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3.
Cell Res., 25, 2015
|
|
1X7N
 
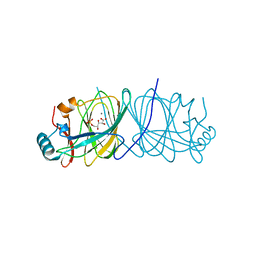 | | The crystal structure of Pyrococcus furiosus phosphoglucose isomerase with bound 5-phospho-D-arabinonate and Manganese | | Descriptor: | 5-PHOSPHOARABINONIC ACID, Glucose-6-phosphate isomerase, MANGANESE (II) ION | | Authors: | Berrisford, J.M, Akerboom, J, Brouns, S, Sedelnikova, S.E, Turnbull, A.P, van der Oost, J, Salmon, L, Hardre, R, Murray, I.A, Blackburn, G.M, Rice, D.W, Baker, P.J. | | Deposit date: | 2004-08-16 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The structures of inhibitor complexes of Pyrococcus furiosus phosphoglucose isomerase provide insights into substrate binding and catalysis.
J.Mol.Biol., 343, 2004
|
|
8P1N
 
 | |
8OPF
 
 | |
8OPK
 
 | |
