4JA9
 
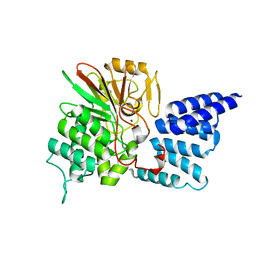 | | Rat PP5 apo | | Descriptor: | MAGNESIUM ION, Serine/threonine-protein phosphatase 5 | | Authors: | Haslbeck, V, Helmuth, M, Alte, F, Popowicz, G, Schmidt, W, Weiwad, M, Fischer, G, Gemmecker, G, Sattler, M, Striggow, F, Groll, M, Richter, K. | | Deposit date: | 2013-02-18 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Selective activators of protein phosphatase 5 target the auto-inhibitory mechanism.
Biosci.Rep., 35, 2015
|
|
4JA7
 
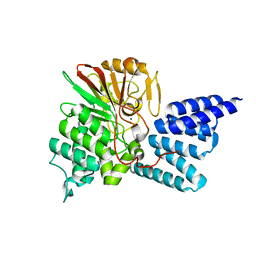 | | Rat PP5 co-crystallized with P5SA-2 | | Descriptor: | MAGNESIUM ION, Serine/threonine-protein phosphatase 5 | | Authors: | Haslbeck, V, Helmuth, M, Alte, F, Popowicz, G, Schmidt, W, Weiwad, M, Fischer, G, Gemmecker, G, Sattler, M, Striggow, F, Groll, M, Richter, K. | | Deposit date: | 2013-02-18 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective activators of protein phosphatase 5 target the auto-inhibitory mechanism.
Biosci.Rep., 35, 2015
|
|
4YZM
 
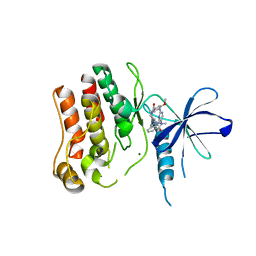 | | Humanized Roco4 bound to LRRK2-In1 | | Descriptor: | 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, MAGNESIUM ION, Probable serine/threonine-protein kinase roco4 | | Authors: | Gilsbach, B.K, Messias, A.C, Ito, G, Sattler, M, Alessi, D.R, Wittinghofer, A, Kortholt, A. | | Deposit date: | 2015-03-25 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Characterization of LRRK2 Inhibitors.
J.Med.Chem., 58, 2015
|
|
5AGQ
 
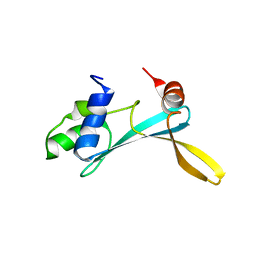 | | Solution structure of the TAM domain of human TIP5 BAZ2A involved in epigenetic regulation of rRNA genes | | Descriptor: | BROMODOMAIN ADJACENT TO ZINC FINGER DOMAIN PROTEIN 2A | | Authors: | Anosova, I, Melnik, S, Tripsianes, K, Kateb, F, Grummt, I, Sattler, M. | | Deposit date: | 2015-02-03 | | Release date: | 2015-05-06 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A Novel RNA Binding Surface of the Tam Domain of Tip5/Baz2A Mediates Epigenetic Regulation of Rrna Genes.
Nucleic Acids Res., 43, 2015
|
|
5JU7
 
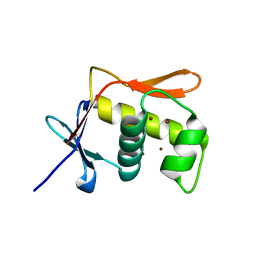 | | DNA BINDING DOMAIN OF E.COLI CADC | | Descriptor: | Transcriptional activator CadC, ZINC ION | | Authors: | Janowski, R, Schlundt, A, Sattler, M, Niessing, D. | | Deposit date: | 2016-05-10 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-function analysis of the DNA-binding domain of a transmembrane transcriptional activator.
Sci Rep, 7, 2017
|
|
1LXL
 
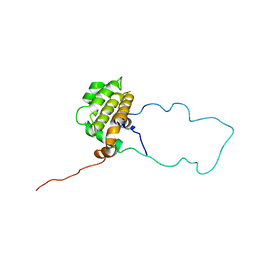 | | NMR STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | BCL-XL | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-04 | | Release date: | 1997-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
6SPT
 
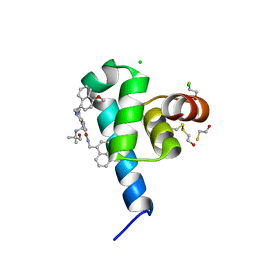 | | High resolution crystal structure of N-terminal domain of PEX14 from Trypanosoma brucei in complex with the fist compound with sub-micromolar trypanocidal activity | | Descriptor: | 5-[(4-methoxynaphthalen-1-yl)methyl]-1-[2-[(2-methyl-1-oxidanyl-propan-2-yl)amino]ethyl]-~{N}-(naphthalen-1-ylmethyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridine-3-carboxamide, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Napolitano, V, Dawidowski, M, Kalel, V.C, Fino, R, Emmanouilidis, L, Lenhart, D, Ostertag, M, Kaiser, M, Kolonko, M, Schilebs, W, Maser, P, Tetko, I, Hadian, K, Plettenburg, O, Erdmann, R, Sattler, M, Popowicz, G.M, Dubin, G. | | Deposit date: | 2019-09-02 | | Release date: | 2020-01-01 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-Activity Relationship in Pyrazolo[4,3-c]pyridines, First Inhibitors of PEX14-PEX5 Protein-Protein Interaction with Trypanocidal Activity.
J.Med.Chem., 63, 2020
|
|
1H5P
 
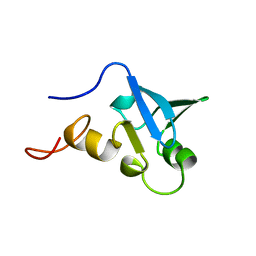 | | Solution structure of the human Sp100b SAND domain by heteronuclear NMR. | | Descriptor: | NUCLEAR AUTOANTIGEN SP100-B | | Authors: | Bottomley, M.J, Liu, Z, Collard, M.W, Huggenvik, J.I, Gibson, T.J, Sattler, M. | | Deposit date: | 2001-05-24 | | Release date: | 2001-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation.
Nat. Struct. Biol., 8, 2001
|
|
1O0P
 
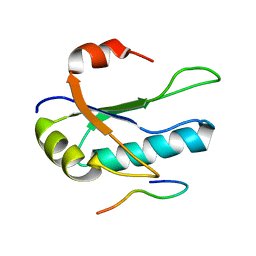 | | Solution Structure of the third RNA Recognition Motif (RRM) of U2AF65 in complex with an N-terminal SF1 peptide | | Descriptor: | Splicing Factor SF1, Splicing factor U2AF 65 kDa subunit | | Authors: | Selenko, P, Gregorovic, G, Sprangers, R, Stier, G, Rhani, Z, Kramer, A, Sattler, M. | | Deposit date: | 2003-02-24 | | Release date: | 2003-05-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP
Mol.Cell, 11, 2003
|
|
1MAZ
 
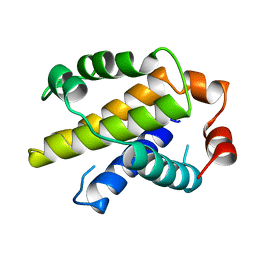 | | X-RAY STRUCTURE OF BCL-XL, AN INHIBITOR OF PROGRAMMED CELL DEATH | | Descriptor: | Bcl-2-like protein 1 | | Authors: | Muchmore, S.W, Sattler, M, Liang, H, Meadows, R.P, Harlan, J.E, Yoon, H.S, Nettesheim, D, Chang, B.S, Thompson, C.B, Wong, S.L, Ng, S.C, Fesik, S.W. | | Deposit date: | 1996-04-09 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death.
Nature, 381, 1996
|
|
5LSO
 
 | |
1G5V
 
 | | SOLUTION STRUCTURE OF THE TUDOR DOMAIN OF THE HUMAN SMN PROTEIN | | Descriptor: | SURVIVAL MOTOR NEURON PROTEIN 1 | | Authors: | Selenko, P, Sprangers, R, Stier, G, Buehler, D, Fischer, U, Sattler, M. | | Deposit date: | 2000-11-02 | | Release date: | 2001-05-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | SMN tudor domain structure and its interaction with the Sm proteins.
Nat.Struct.Biol., 8, 2001
|
|
1OPI
 
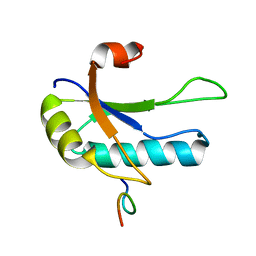 | | SOLUTION STRUCTURE OF THE THIRD RNA RECOGNITION MOTIF (RRM) OF U2AF65 IN COMPLEX WITH AN N-TERMINAL SF1 PEPTIDE | | Descriptor: | SPLICING FACTOR SF1, SPLICING FACTOR U2AF 65 KDA SUBUNIT | | Authors: | Selenko, P, Gregorovic, G, Sprangers, R, Stier, G, Rhani, Z, Kramer, A, Sattler, M. | | Deposit date: | 2003-03-05 | | Release date: | 2004-03-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP
Mol.Cell, 11, 2003
|
|
7PO6
 
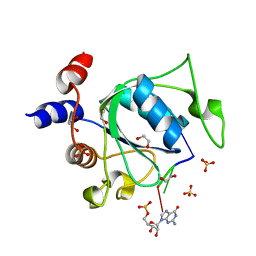 | | Xist (m6A)UCG tetraloop RNA bound to the YTH domain of YTHDC1 | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-TRIPHOSPHATE, Isoform 2 of YTH domain-containing protein 1, ... | | Authors: | Jones, A.N, Mourao, A, Sattler, M. | | Deposit date: | 2021-09-08 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural effects of m6A modification of the Xist A-repeat AUCG tetraloop and its recognition by YTHDC1.
Nucleic Acids Res., 50, 2022
|
|
5MMC
 
 | |
6I5P
 
 | |
5N8V
 
 | | Targeting the PEX14-PEX5 interaction by small molecules provides novel therapeutic routes to treat trypanosomiases. | | Descriptor: | 1-(2-azanylethyl)-5-[(4-methoxynaphthalen-1-yl)methyl]-~{N}-(naphthalen-1-ylmethyl)-6,7-dihydro-4~{H}-pyrazolo[4,3-c]pyridine-3-carboxamide, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Dawidowski, M, Emmanouilidis, L, Sattler, M, Popowicz, G.M. | | Deposit date: | 2017-02-24 | | Release date: | 2017-03-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Inhibitors of PEX14 disrupt protein import into glycosomes and kill Trypanosoma parasites.
Science, 355, 2017
|
|
4JVU
 
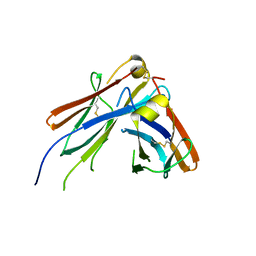 | | IgM C2-domain from mouse | | Descriptor: | Ig mu chain C region membrane-bound form | | Authors: | Mueller, R, Graewert, A.M, Kern, T, Madl, T, Peschek, J, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2013-03-26 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6I7A
 
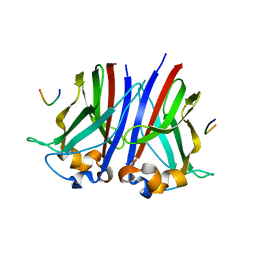 | |
6I68
 
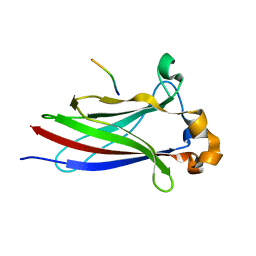 | |
4JVW
 
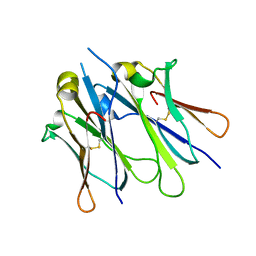 | | IgM C4-domain from mouse | | Descriptor: | Ig mu chain C region secreted form | | Authors: | Mueller, R, Graewert, A.M, Kern, T, Madl, T, Peschek, J, Sattler, M, Groll, M, Buchner, J. | | Deposit date: | 2013-03-26 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of the IgM Fc domains reveal principles of its hexamer formation.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6I41
 
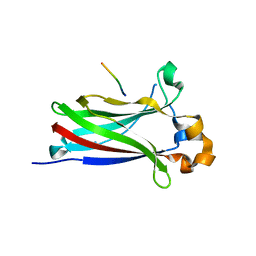 | |
1ZZP
 
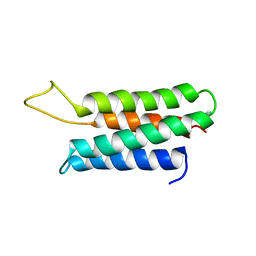 | | Solution structure of the F-actin binding domain of Bcr-Abl/c-Abl | | Descriptor: | Proto-oncogene tyrosine-protein kinase ABL1 | | Authors: | Hantschel, O, Wiesner, S, Guttler, T, Mackereth, C.D, Rix, L.L.R, Mikes, Z, Dehne, J, Gorlich, D, Sattler, M, Superti-Furga, G. | | Deposit date: | 2005-06-14 | | Release date: | 2005-08-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Cytoskeletal Association of Bcr-Abl/c-Abl.
Mol.Cell, 19, 2005
|
|
6R73
 
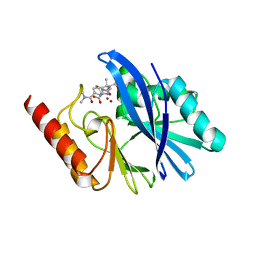 | | Structure of IMP-13 metallo-beta-lactamase complexed with hydrolysed meropenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, Beta-lactamase, ZINC ION | | Authors: | Softley, C.A, Zak, K, Kolonko, M, Sattler, M, Popowicz, G. | | Deposit date: | 2019-03-28 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
6R78
 
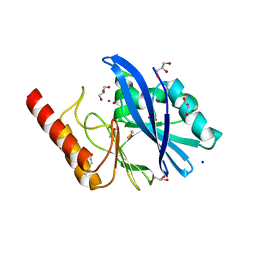 | | Structure of IMP-13 metallo-beta-lactamase in apo form (loop closed) | | Descriptor: | 1,2-ETHANEDIOL, BETA-MERCAPTOETHANOL, Beta-lactamase, ... | | Authors: | Zak, K.M, Softley, C, Kolonko, M, Sattler, M, Popowicz, G.M. | | Deposit date: | 2019-03-28 | | Release date: | 2020-04-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structure and Molecular Recognition Mechanism of IMP-13 Metallo-beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2020
|
|
