6IWR
 
 | | Crystal structure of GalNAc-T7 with UDP, GalNAc and Mn2+ | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MANGANESE (II) ION, N-acetylgalactosaminyltransferase 7, ... | | Authors: | Yu, C, Yin, Y.X. | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structural basis of carbohydrate transfer activity of UDP-GalNAc: Polypeptide N-acetylgalactosaminyltransferase 7.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
6IWQ
 
 | | Crystal structure of GalNAc-T7 with Mn2+ | | Descriptor: | MANGANESE (II) ION, N-acetylgalactosaminyltransferase 7 | | Authors: | Yu, C, Yin, Y.X. | | Deposit date: | 2018-12-06 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis of carbohydrate transfer activity of UDP-GalNAc: Polypeptide N-acetylgalactosaminyltransferase 7.
Biochem. Biophys. Res. Commun., 510, 2019
|
|
6JXT
 
 | |
6JX0
 
 | |
6IGO
 
 | | Crystal structure of myelin protein zero-like protein 1 (MPZL1) | | Descriptor: | Myelin protein zero-like protein 1 | | Authors: | Yu, T. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Structural and biochemical studies of the extracellular domain of Myelin protein zero-like protein 1
Biochem. Biophys. Res. Commun., 506, 2018
|
|
6JWL
 
 | | Crystal structure of EGFR 696-1022 L858R in complex with AZD9291 | | Descriptor: | Epidermal growth factor receptor, N-(2-{[2-(dimethylamino)ethyl](methyl)amino}-4-methoxy-5-{[4-(1-methyl-1H-indol-3-yl)pyrimidin-2-yl]amino}phenyl)prop-2-enamide | | Authors: | Yun, C.H, Zhu, S.J, Yan, X.E. | | Deposit date: | 2019-04-21 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Structural Basis of AZD9291 Selectivity for EGFR T790M.
J.Med.Chem., 63, 2020
|
|
6JX4
 
 | |
6IGT
 
 | | MPZL1 mutant - V145G, Q146K, P147T and G148S | | Descriptor: | Myelin protein zero-like protein 1 | | Authors: | Yu, T. | | Deposit date: | 2018-09-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Structural and biochemical studies of the extracellular domain of Myelin protein zero-like protein 1
Biochem. Biophys. Res. Commun., 506, 2018
|
|
6IGW
 
 | | MPZL1 mutant - S86G, V145G, Q146K,P147T,G148S | | Descriptor: | Myelin protein zero-like protein 1 | | Authors: | Yu, T. | | Deposit date: | 2018-09-26 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Structural and biochemical studies of the extracellular domain of Myelin protein zero-like protein 1
Biochem. Biophys. Res. Commun., 506, 2018
|
|
8GRQ
 
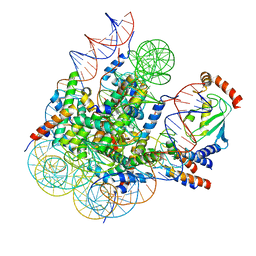 | | Cryo-EM structure of BRCA1/BARD1 bound to H2AK127-UbcH5c-Ub nucleosome | | Descriptor: | BRCA1-associated RING domain protein 1, Breast cancer type 1 susceptibility protein, DNA (147-MER), ... | | Authors: | Ai, H.S, Zebin, T, Zhiheng, D, Jiakun, T, Liying, Z, Jia-Bin, L, Man, P, Liu, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.87 Å) | | Cite: | Synthetic E2-Ub-nucleosome conjugates for studying nucleosome ubiquitination.
Chem, 2023
|
|
8GRM
 
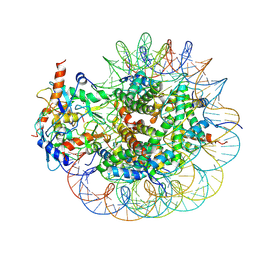 | | Cryo-EM structure of PRC1 bound to H2AK119-UbcH5b-Ub nucleosome | | Descriptor: | COMMD3 protein, DNA (144-MER), DNA (145-MER), ... | | Authors: | Ai, H.S, Zebin, T, Zhihend, D, Jiakun, T, Liying, Z, Jia-Bin, L, Man, P, Liu, L. | | Deposit date: | 2022-09-02 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Synthetic E2-Ub-nucleosome conjugates for studying nucleosome ubiquitination.
Chem, 2023
|
|
7XCT
 
 | | Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 2:1 complex | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-04-20 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XD1
 
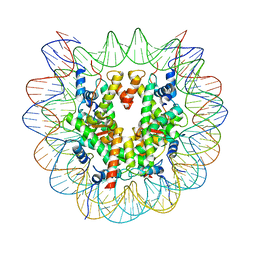 | | cryo-EM structure of unmodified nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-26 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XD0
 
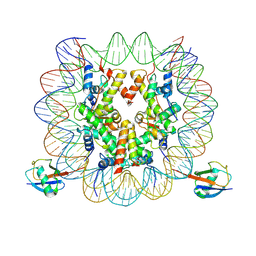 | | cryo-EM structure of H2BK34ub nucleosome | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-26 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XCR
 
 | | Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 1:1 complex | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-04-20 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
8HQY
 
 | | Cryo-EM structure of SSX1 bound to the H2AK119Ub nucleosome at a resolution of 3.05 angstrom | | Descriptor: | DNA (136-MER), DNA (137-MER), Histone H2A type 1-B/E, ... | | Authors: | Zebin, T, Ai, H.S, Ziyu, X, GuoChao, C, Man, P, Liu, L. | | Deposit date: | 2022-12-14 | | Release date: | 2023-09-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Synovial sarcoma X breakpoint 1 protein uses a cryptic groove to selectively recognize H2AK119Ub nucleosomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8HR1
 
 | | Cryo-EM structure of SSX1 bound to the unmodified nucleosome at a resolution of 3.02 angstrom | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Zebin, T, Ai, H.S, Ziyu, X, Man, P, Liu, L. | | Deposit date: | 2022-12-14 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Synovial sarcoma X breakpoint 1 protein uses a cryptic groove to selectively recognize H2AK119Ub nucleosomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7UP2
 
 | | NDM1-inhibitor co-structure | | Descriptor: | (2M)-4'-methyl-2-(2H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UP1
 
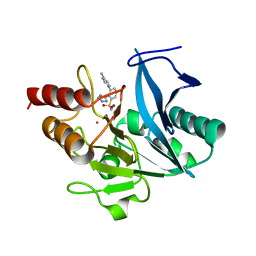 | | NDM1-inhibitor co-structure | | Descriptor: | (2M)-4'-methyl-2-(2H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, CADMIUM ION, Metallo beta-lactamase, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UP3
 
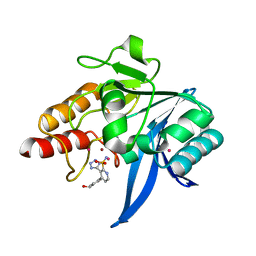 | | NDM1-inhibitor co-structure | | Descriptor: | (3P)-4-[4-(hydroxymethyl)phenyl]-3-(2H-tetrazol-5-yl)pyridine-2-sulfonamide, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CADMIUM ION, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UOX
 
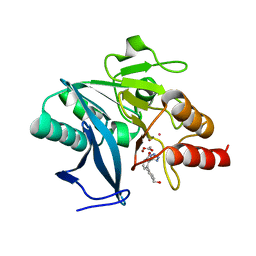 | | NDM1-inhibitor co-structure | | Descriptor: | (2M)-4'-(hydroxymethyl)-2-(1H-tetrazol-5-yl)[1,1'-biphenyl]-3-ol, ACETATE ION, CADMIUM ION, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
7UOY
 
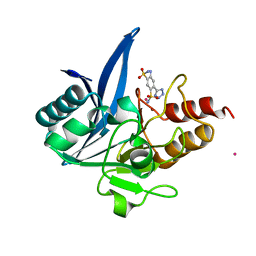 | | NDM1-inhibitor co-structure | | Descriptor: | (6P)-4-amino-6-(2H-tetrazol-5-yl)benzene-1,3-disulfonamide, CADMIUM ION, Metallo beta-lactamase, ... | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
4MQ1
 
 | | The crystal structure of DYRK1a with a bound pyrido[2,3-d]pyrimidine inhibitor | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 1A, N-(5-{[(1R)-3-amino-1-(3-chlorophenyl)propyl]carbamoyl}-2-chlorophenyl)-2-methoxy-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidine-6-carboxamide, PENTAETHYLENE GLYCOL, ... | | Authors: | Lukacs, C.M, Janson, C.A, Garvie, C, Liang, L. | | Deposit date: | 2013-09-15 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Pyrido[2,3-d]pyrimidines: Discovery and preliminary SAR of a novel series of DYRK1B and DYRK1A inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7UGW
 
 | | M. tuberculosis DNA gyrase cleavage core bound to DNA and evybactin | | Descriptor: | DNA (46-MER), DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Hauk, G, Imai, Y, Lewis, K, Berger, J.M. | | Deposit date: | 2022-03-25 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Evybactin is a DNA gyrase inhibitor that selectively kills Mycobacterium tuberculosis.
Nat.Chem.Biol., 18, 2022
|
|
