5HLW
 
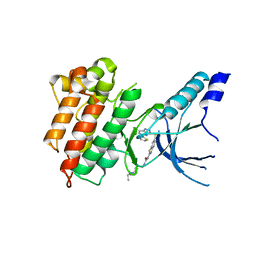 | | Crystal structure of c-Met mutant Y1230H in complex with compound 14 | | Descriptor: | 1-[2-(1-ethylpiperidin-4-yl)ethyl]-3-(6-{[6-(thiophen-2-yl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)urea, CHLORIDE ION, Hepatocyte growth factor receptor | | Authors: | Vallee, F, Pouzieux, S, Marquette, J.P, Houtmann, J. | | Deposit date: | 2016-01-15 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
5HOA
 
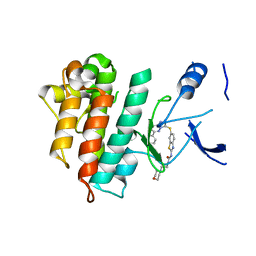 | | Crystal structure of c-Met L1195V in complex with SAR125844 | | Descriptor: | 1-(6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)-3-[2-(morpholin-4-yl)ethyl]urea, Hepatocyte growth factor receptor | | Authors: | Vallee, F, Marquette, J.-P. | | Deposit date: | 2016-01-19 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
6NT2
 
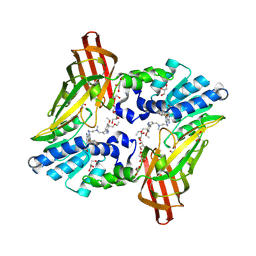 | | type 1 PRMT in complex with the inhibitor GSK3368715 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, N~1~-({5-[4,4-bis(ethoxymethyl)cyclohexyl]-1H-pyrazol-4-yl}methyl)-N~1~,N~2~-dimethylethane-1,2-diamine, ... | | Authors: | Concha, N.O. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Anti-tumor Activity of the Type I PRMT Inhibitor, GSK3368715, Synergizes with PRMT5 Inhibition through MTAP Loss.
Cancer Cell, 36, 2019
|
|
7AYZ
 
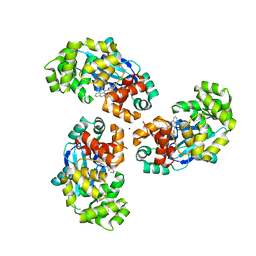 | |
7AZ0
 
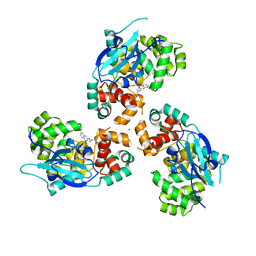 | |
7AYY
 
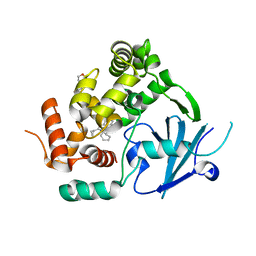 | | Structure of the human 8-oxoguanine DNA Glycosylase hOGG1 in complex with activator TH10785 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Davies, J.R, Stenmark, P. | | Deposit date: | 2020-11-13 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Small-molecule activation of OGG1 increases oxidative DNA damage repair by gaining a new function.
Science, 376, 2022
|
|
5HNI
 
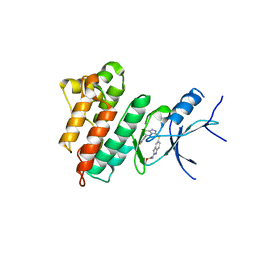 | | CRYSTAL STRUCTURE OF CMET WT with compound 3 | | Descriptor: | Hepatocyte growth factor receptor, methyl (6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1H-benzimidazol-2-yl)carbamate | | Authors: | Vallee, F, Houtmann, J. | | Deposit date: | 2016-01-18 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
5HO6
 
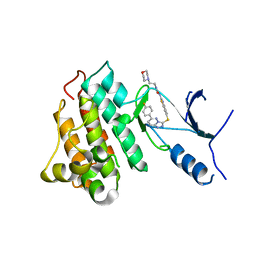 | | CRYSTAL STRUCTURE OF CMET IN COMPLEX WITH CMPD. | | Descriptor: | 1-(6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)-3-[2-(morpholin-4-yl)ethyl]urea, Hepatocyte growth factor receptor | | Authors: | Vallee, F, Houtmann, J. | | Deposit date: | 2016-01-19 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
6G3X
 
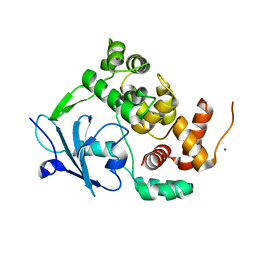 | |
6VER
 
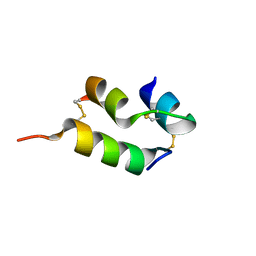 | | Human insulin analog: [GluB10,TyrB20]-DOI | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Menting, J.G, Chou, D.H.-C, Lawrence, M.C, Xiong, X. | | Deposit date: | 2020-01-02 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.047 Å) | | Cite: | Mini-Ins: A minimal, bioactive insulin analog with alternative binding modes
not published
|
|
6G3Y
 
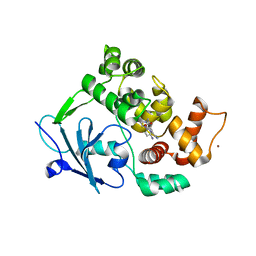 | | Structure of the mouse 8-oxoguanine DNA Glycosylase mOGG1 in complex with ligand TH5675 | | Descriptor: | 4-(4-azanyl-2-oxidanylidene-3~{H}-benzimidazol-1-yl)-~{N}-(4-iodophenyl)piperidine-1-carboxamide, ACETATE ION, N-glycosylase/DNA lyase, ... | | Authors: | Masuyer, G, Helleday, T, Stenmark, P. | | Deposit date: | 2018-03-26 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Small-molecule inhibitor of OGG1 suppresses proinflammatory gene expression and inflammation.
Science, 362, 2018
|
|
6VES
 
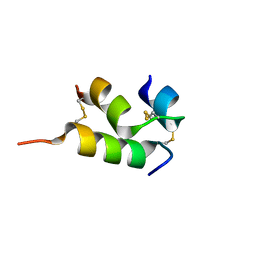 | | Human insulin analog: [GluB10,HisA8,ArgA9]-DOI | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Menting, J.G, Chou, D.H.-C, Lawrence, M.C, Xiong, X. | | Deposit date: | 2020-01-02 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mini-Ins: A Minimal, Bioactive Insulin Analog with Alternative Binding Modes
To Be Published
|
|
7KGB
 
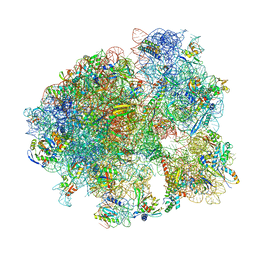 | |
3U9Z
 
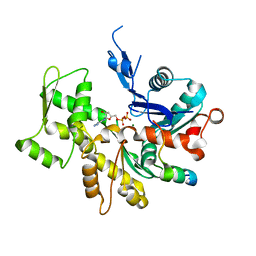 | | Crystal structure between actin and a protein construct containing the first beta-thymosin domain of drosophila ciboulot (residues 2-58) with the three mutations N26D/Q27K/D28S | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-10-20 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly
Embo J., 31, 2012
|
|
3U9D
 
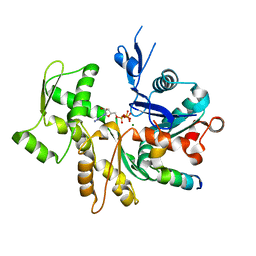 | | Crystal Structure of a chimera containing the N-terminal domain (residues 8-24) of drosophila Ciboulot and the C-terminal domain (residues 13-44) of bovine Thymosin-beta4, bound to G-actin-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-10-18 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly.
Embo J., 31, 2012
|
|
3U8X
 
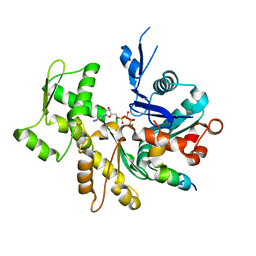 | | Crystal Structure of a chimera containing the N-terminal domain (residues 8-29) of drosophila Ciboulot and the C-terminal domain (residues 18-44) of bovine Thymosin-beta4, bound to G-actin-ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Renault, L, Husson, C, Carlier, M.F, Didry, D. | | Deposit date: | 2011-10-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly
Embo J., 31, 2012
|
|
7AZS
 
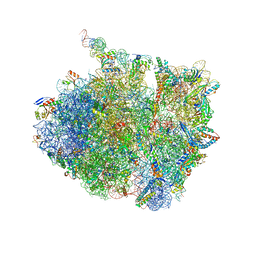 | | 70S thermus thermophilus ribosome with bound antibiotic lead SEQ-569 | | Descriptor: | (2R,3S,4R,5R,7S,9S,10S,11R,12S,13R)-12-(((2R,4R,5S,6S)-4,5-dihydroxy-4,6-dimethyltetrahydro-2H-pyran-2-yl)oxy)-2-((S)-1-(((2R,3R,4R,5R,6R)-5-hydroxy-3,4-dimethoxy-6-methyltetrahydro-2H-pyran-2-yl)oxy)propan-2-yl)-10-(((2S,3R,6R,E)-3-hydroxy-4-(methoxyimino)-6-methyltetrahydro-2H-pyran-2-yl)oxy)-3,5,7,9,11,13-hexamethyl-7-(((2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethyl)carbamoyl)oxy)-6,14-dioxooxacyclotetradecan-4-yl 3-methylbutanoate, 16S rRNA, 23S rRNA, ... | | Authors: | Jenner, L.B, Yusupov, M, Yusupova, G, Rak, A. | | Deposit date: | 2020-11-17 | | Release date: | 2022-06-08 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of natural-product-derived sequanamycins as potent oral anti-tuberculosis agents.
Cell, 186, 2023
|
|
7AZO
 
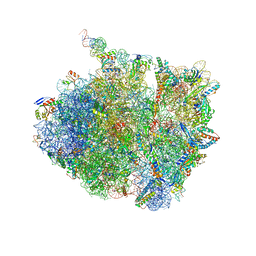 | | 70S thermus thermophilus ribosome with bound antibiotic lead SEQ-977 | | Descriptor: | (2R,3S,4R,5R,7S,9S,10S,11R,12S,13R)-12-(((2R,4R,5S,6S)-4,5-dihydroxy-4,6-dimethyltetrahydro-2H-pyran-2-yl)oxy)-2-((S)-1-(((2R,3R,4R,5R,6R)-5-hydroxy-3,4-dimethoxy-6-methyltetrahydro-2H-pyran-2-yl)oxy)propan-2-yl)-10-(((2S,3R,4R,6R)-3-hydroxy-4-(methoxyamino)-6-methyltetrahydro-2H-pyran-2-yl)oxy)-3,5,7,9,11,13-hexamethyl-7-(((2-((2-nitrophenyl)sulfonamido)ethyl)carbamoyl)oxy)-6,14-dioxooxacyclotetradecan-4-yl 3-methylbutanoate, 16S rRNA, 23S rRNA, ... | | Authors: | Jenner, L.B, Yusupov, M, Yusupova, G. | | Deposit date: | 2020-11-17 | | Release date: | 2022-06-01 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Discovery of natural-product-derived sequanamycins as potent oral anti-tuberculosis agents.
Cell, 186, 2023
|
|
6XOG
 
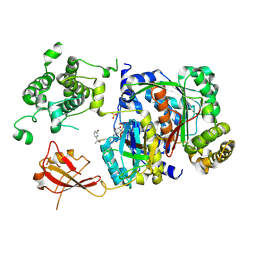 | | Structure of SUMO1-ML786519 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XOH
 
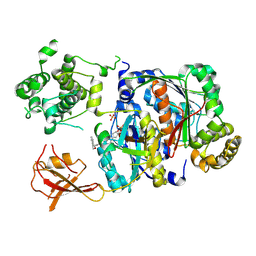 | | Structure of SUMO1-ML00789344 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.226 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
6XOI
 
 | | Structure of SUMO1-ML00752641 adduct bound to SAE | | Descriptor: | SULFATE ION, SUMO-activating enzyme subunit 1, SUMO-activating enzyme subunit 2, ... | | Authors: | Sintchak, M, Lane, W, Bump, N. | | Deposit date: | 2020-07-07 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of TAK-981, a First-in-Class Inhibitor of SUMO-Activating Enzyme for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
4MTY
 
 | | Structure at 1A resolution of a helical aromatic foldamer-protein complex. | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ogayone, T, Buratto, J, Langlois D'Estaintot, B, Stupfel, M, Granier, T, Gallois, B, Huc, Y. | | Deposit date: | 2013-09-20 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of a complex formed by a protein and a helical aromatic oligoamide foldamer at 2.1 a resolution.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4LP6
 
 | | Crystal Structure of Human Carbonic Anhydrase II in complex with a quinoline oligoamide foldamer | | Descriptor: | 8-({[4-(3-aminopropoxy)-8-({[4-hydroxy-8-({[4-(2-methylpropoxy)-8-({[4-(3-{[(4-sulfamoylbenzoyl)amino]methyl}phenoxy)butyl]carbamoyl}amino)quinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)-4-(carboxymethoxy)quinoline-2-carboxylic acid, Carbonic anhydrase 2, ZINC ION | | Authors: | Buratto, J, Granier, T, Langlois D'estaintot, B, Huc, I, Gallois, B. | | Deposit date: | 2013-07-15 | | Release date: | 2013-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of a complex formed by a protein and a helical aromatic oligoamide foldamer at 2.1 angstrom resolution.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6ARJ
 
 | | Crystal structure of CARM1 with EPZ022302 and SAH | | Descriptor: | GLYCEROL, Histone-arginine methyltransferase CARM1, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Boriack-Sjodin, P.A, Jin, L. | | Deposit date: | 2017-08-22 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of a CARM1 Inhibitor with Potent In Vitro and In Vivo Activity in Preclinical Models of Multiple Myeloma.
Sci Rep, 7, 2017
|
|
6ARV
 
 | | Crystal structure of CARM1 with Compound 2 and SAH | | Descriptor: | (2R)-1-amino-3-{3-[4-(morpholin-4-yl)-1-(propan-2-yl)-1H-pyrazolo[3,4-b]pyridin-6-yl]phenoxy}propan-2-ol, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Boriack-Sjodin, P.A, Jin, L. | | Deposit date: | 2017-08-23 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a CARM1 Inhibitor with Potent In Vitro and In Vivo Activity in Preclinical Models of Multiple Myeloma.
Sci Rep, 7, 2017
|
|
