5O5B
 
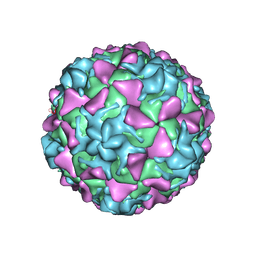 | | Poliovirus type 3 (strain Saukett) stabilized virus-like particle | | Descriptor: | Capsid proteins, VP1, VP2, ... | | Authors: | Bahar, M.W, Kotecha, A, Fry, E.E, Stuart, D.I. | | Deposit date: | 2017-06-01 | | Release date: | 2017-07-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Plant-made polio type 3 stabilized VLPs-a candidate synthetic polio vaccine.
Nat Commun, 8, 2017
|
|
5O5P
 
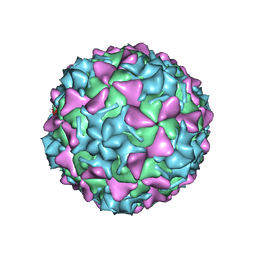 | | Poliovirus type 3 (strain Saukett) stabilized virus-like particle in complex with the pocket factor compound GPP3 | | Descriptor: | 1-[5-[4-(ethoxyiminomethyl)phenoxy]-3-methyl-pentyl]-3-pyridin-4-yl-imidazol-2-one, Capsid proteins, VP4, ... | | Authors: | Bahar, M.W, Kotecha, A, Fry, E.E, Stuart, D.I. | | Deposit date: | 2017-06-02 | | Release date: | 2017-07-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Plant-made polio type 3 stabilized VLPs-a candidate synthetic polio vaccine.
Nat Commun, 8, 2017
|
|
8ANW
 
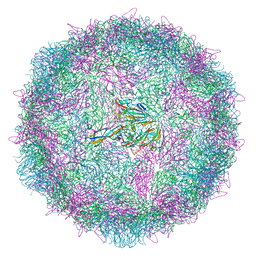 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8). | | Descriptor: | Capsid protein, VP0, VP1, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-08-06 | | Release date: | 2022-10-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Production and Characterisation of Stabilised PV-3 Virus-like Particles Using Pichia pastoris .
Viruses, 14, 2022
|
|
8AYY
 
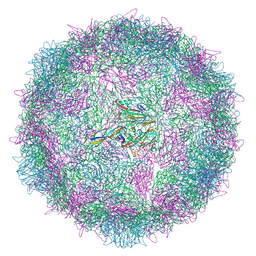 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and Pleconaril | | Descriptor: | 3-{3,5-DIMETHYL-4-[3-(3-METHYL-ISOXAZOL-5-YL)-PROPOXY]-PHENYL}-5-TRIFLUOROMETHYL-[1,2,4]OXADIAZOLE, Capsid protein, VP0, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-09-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A conserved glutathione binding site in poliovirus is a target for antivirals and vaccine stabilisation.
Commun Biol, 5, 2022
|
|
8AYX
 
 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8) in complex with GSH and GPP3 | | Descriptor: | 1-[(3S)-5-[4-[(E)-ETHOXYIMINOMETHYL]PHENOXY]-3-METHYL-PENTYL]-3-PYRIDIN-4-YL-IMIDAZOLIDIN-2-ONE, Capsid protein, VP0, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-09-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A conserved glutathione binding site in poliovirus is a target for antivirals and vaccine stabilisation.
Commun Biol, 5, 2022
|
|
8AYZ
 
 | | Poliovirus type 2 (strain MEF-1) virus-like particle in complex with capsid binder compound 17 | | Descriptor: | 4-[[4-[1,3-bis(oxidanylidene)isoindol-2-yl]phenyl]sulfonylamino]benzoic acid, Capsid protein, VP0, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-09-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (1.88 Å) | | Cite: | A conserved glutathione binding site in poliovirus is a target for antivirals and vaccine stabilisation.
Commun Biol, 5, 2022
|
|
7QNY
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with COVOX-58 and COVOX-158 Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-158 heavy chain, COVOX-158 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
1BBT
 
 | | METHODS USED IN THE STRUCTURE DETERMINATION OF FOOT AND MOUTH DISEASE VIRUS | | Descriptor: | FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP1), FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP2), FOOT-AND-MOUTH DISEASE VIRUS (SUBUNIT VP3), ... | | Authors: | Acharya, K.R, Fry, E.E, Logan, D.T, Stuart, D.I. | | Deposit date: | 1992-05-18 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Methods used in the structure determination of foot-and-mouth disease virus.
Acta Crystallogr.,Sect.A, 49, 1993
|
|
7QNW
 
 | | The receptor binding domain of SARS-CoV-2 Omicron variant spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Beta-55 heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
7QNX
 
 | | The receptor binding domain of SARS-CoV-2 spike glycoprotein in complex with Beta-55 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-55 heavy chain, Beta-55 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2021-12-23 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses.
Cell, 185, 2022
|
|
9F9Y
 
 | | SARS-CoV-2 BA-2.87.1 Spike ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ren, J, Stuart, D.I, Duyvesteyn, H.M.E. | | Deposit date: | 2024-05-09 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Concerted deletions eliminate a neutralizing supersite in SARS-CoV-2 BA.2.87.1 spike.
Structure, 32, 2024
|
|
5OYI
 
 | | FMDV A10 dissociated pentamer | | Descriptor: | Genome polyprotein | | Authors: | Kotecha, A, Malik, N, Stuart, D.I. | | Deposit date: | 2017-09-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Structures of foot and mouth disease virus pentamers: Insight into capsid dissociation and unexpected pentamer reassociation.
PLoS Pathog., 13, 2017
|
|
4Q4F
 
 | | Crystal structure of LIMP-2 (space group C2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-beta-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Y, Ren, J, Padilla-Parra, S, Fry, L.E, Stuart, D.I. | | Deposit date: | 2014-04-14 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lysosome sorting of beta-glucocerebrosidase by LIMP-2 is targeted by the mannose 6-phosphate receptor.
Nat Commun, 5, 2014
|
|
4Q4B
 
 | | Crystal structure of LIMP-2 (space group C2221) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosome membrane protein 2, ... | | Authors: | Zhao, Y, Ren, J, Padilla-Parra, S, Fry, L.E, Stuart, D.I. | | Deposit date: | 2014-04-14 | | Release date: | 2014-07-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Lysosome sorting of beta-glucocerebrosidase by LIMP-2 is targeted by the mannose 6-phosphate receptor.
Nat Commun, 5, 2014
|
|
7NX6
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab Heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-03-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NXA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 B.1.351 variant Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-222 Fab heavy chain, COVOX-222 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-03-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NXB
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 P.1 variant Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-222 Fab heavy chain, COVOX-222 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-03-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NXC
 
 | |
7NX9
 
 | | Crystal structure of the N501Y mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-03-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NX7
 
 | | Crystal structure of the K417N mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-03-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NX8
 
 | | Crystal structure of the K417T mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-03-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
5JQ3
 
 | | Crystal structure of Ebola glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein 1,Envelope glycoprotein 1,Envelope glycoprotein 1, Envelope glycoprotein 2, ... | | Authors: | Zhao, Y, Ren, J, Stuart, D.I. | | Deposit date: | 2016-05-04 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Toremifene interacts with and destabilizes the Ebola virus glycoprotein.
Nature, 535, 2016
|
|
5JQB
 
 | | Crystal structure of Ebola glycoprotein in complex with ibuprofen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein 1,Envelope glycoprotein 1,Envelope glycoprotein 1, Envelope glycoprotein 2, ... | | Authors: | Zhao, Y, Ren, J, Stuart, D.I. | | Deposit date: | 2016-05-04 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Toremifene interacts with and destabilizes the Ebola virus glycoprotein.
Nature, 535, 2016
|
|
5JQ7
 
 | | Crystal structure of Ebola glycoprotein in complex with toremifene | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, Envelope glycoprotein 1,Envelope glycoprotein 1,Envelope glycoprotein 1, ... | | Authors: | Zhao, Y, Ren, J, Stuart, D.I. | | Deposit date: | 2016-05-04 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Toremifene interacts with and destabilizes the Ebola virus glycoprotein.
Nature, 535, 2016
|
|
6HS4
 
 | | Crystal structure of Ebolavirus glycoprotein in complex with inhibitor 118 | | Descriptor: | 1-[2-[3-oxidanyl-4-(4-phenyl-1~{H}-pyrazol-5-yl)phenoxy]ethyl]piperidine-4-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Ren, J, Zhao, Y, Stuart, D.I. | | Deposit date: | 2018-09-28 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Based in Silico Screening Identifies a Potent Ebolavirus Inhibitor from a Traditional Chinese Medicine Library.
J.Med.Chem., 62, 2019
|
|
