6F9K
 
 | |
3U9L
 
 | | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase (NADPH) from Sinorhizobium meliloti | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-19 | | Release date: | 2011-11-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase (NADPH) from Sinorhizobium meliloti
TO BE PUBLISHED
|
|
6SLD
 
 | | Structure of the Phosphatidylcholine Binding Mutant of Yeast Sec14 Homolog Sfh1 (S175I,T177I) in Complex with Phosphatidylinositol | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CRAL-TRIO domain-containing protein YKL091C | | Authors: | Berger, J, Fitz, M, Johnen, P, Shanmugaratnam, S, Hocker, B, Stiel, A.C, Schaaf, G. | | Deposit date: | 2019-08-19 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Phosphatidylcholine Binding Mutant of Yeast Sec14 Homolog Sfh1 (S175I,T177I) in Complex with Phosphatidylinositol
To Be Published
|
|
7VXR
 
 | | Crystal structure of BPSL1038 from Burkholderia pseudomallei | | Descriptor: | BPSL1038, SODIUM ION | | Authors: | Shaibullah, S, Mohd-Sharif, M, Ho, K.L, Firdaus-Raih, M, Nathan, S, Mohamed, R, Teh, A.K, Waterman, J, Ng, C.L. | | Deposit date: | 2021-11-13 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and functional analyses of Burkholderia pseudomallei BPSL1038 reveal a Cas-2/VapD nuclease sub-family.
Commun Biol, 6, 2023
|
|
7VXT
 
 | | Crystal structure of a selenomethionine-labeled BPSL1038 from Burkholderia pseudomallei | | Descriptor: | BETA-MERCAPTOETHANOL, BPSL1038, SODIUM ION | | Authors: | Shaibullah, S, Mohd-Sharif, M, Ho, K.L, Firdaus-Raih, M, Nathan, S, Mohamed, R, Teh, A.K, Waterman, J, Ng, C.L. | | Deposit date: | 2021-11-13 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and functional analyses of Burkholderia pseudomallei BPSL1038 reveal a Cas-2/VapD nuclease sub-family.
Commun Biol, 6, 2023
|
|
3UXY
 
 | | The crystal structure of short chain dehydrogenase from Rhodobacter sphaeroides | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Short-chain dehydrogenase/reductase SDR | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-05 | | Release date: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | The crystal structure of short chain dehydrogenase from Rhodobacter sphaeroides
To be Published
|
|
3UOE
 
 | | The crystal structure of dehydrogenase from Sinorhizobium meliloti | | Descriptor: | Dehydrogenase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-16 | | Release date: | 2011-11-30 | | Method: | X-RAY DIFFRACTION (2.311 Å) | | Cite: | The crystal structure of dehydrogenase from Sinorhizobium meliloti
To be Published
|
|
5H2U
 
 | | Crystal structure of PTK6 Kinase Domain complexed with Dasatinib | | Descriptor: | GLYCEROL, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE, Protein-tyrosine kinase 6 | | Authors: | Thakur, M.K, Birudukota, S, Swaminathan, S, Tyagi, R, Gosu, R. | | Deposit date: | 2016-10-18 | | Release date: | 2017-01-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Co-crystal structures of PTK6: With Dasatinib at 2.24 angstrom , with novel imidazo[1,2-a]pyrazin-8-amine derivative inhibitor at 1.70 angstrom resolution
Biochem. Biophys. Res. Commun., 482, 2017
|
|
3U5T
 
 | | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase from Sinorhizobium meliloti | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-11 | | Release date: | 2011-11-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of 3-oxoacyl-[acyl-carrier-protein] reductase from Sinorhizobium meliloti
To be Published
|
|
3UBO
 
 | | The crystal structure of adenosine kinase from Sinorhizobium meliloti | | Descriptor: | ADENOSINE, ADENOSINE-5'-DIPHOSPHATE, adenosine kinase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-10-24 | | Release date: | 2011-11-09 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | The crystal structure of adenosine kinase from Sinorhizobium meliloti
To be Published
|
|
3OMH
 
 | | Crystal structure of PTPN22 in complex with SKAP-HOM pTyr75 peptide | | Descriptor: | Src kinase-associated phosphoprotein 2, Tyrosine-protein phosphatase non-receptor type 22 | | Authors: | Yu, X, Sun, J.-P, Zhang, S, Zhang, Z.-Y. | | Deposit date: | 2010-08-26 | | Release date: | 2011-06-29 | | Last modified: | 2011-09-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Substrate Specificity of Lymphoid-specific Tyrosine Phosphatase (Lyp) and Identification of Src Kinase-associated Protein of 55 kDa Homolog (SKAP-HOM) as a Lyp Substrate.
J.Biol.Chem., 286, 2011
|
|
3IZD
 
 | | Model of the large subunit RNA expansion segment ES27L-out based on a 6.1 A cryo-EM map of Saccharomyces cerevisiae translating 80S ribosome. 3IZD is a small part (an expansion segment) which is in an alternative conformation to what is in already 3IZF. | | Descriptor: | rRNA expansion segment ES27L in an "out" conformation | | Authors: | Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Becker, T, Bhushan, S, Jossinet, F, Habeck, M, Dindar, G, Franckenberg, S, Marquez, V, Mielke, T, Thomm, M, Berninghausen, O, Beatrix, B, Soeding, J, Westhof, E, Wilson, D.N, Beckmann, R. | | Deposit date: | 2010-10-13 | | Release date: | 2010-12-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Cryo-EM structure and rRNA model of a translating eukaryotic 80S ribosome at 5.5-A resolution.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3V5N
 
 | | The crystal structure of oxidoreductase from Sinorhizobium meliloti | | Descriptor: | Oxidoreductase | | Authors: | Zhang, Z, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-12-16 | | Release date: | 2012-01-04 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | The crystal structure of oxidoreductase from Sinorhizobium meliloti
To be Published
|
|
7T91
 
 | |
3S6J
 
 | | The crystal structure of a hydrolase from Pseudomonas syringae | | Descriptor: | CALCIUM ION, Hydrolase, haloacid dehalogenase-like family | | Authors: | Zhang, Z, Syed Ibrahim, B, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-05-25 | | Release date: | 2011-07-13 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | The crystal structure of a hydrolase from Pseudomonas syringae
To be Published
|
|
3L4A
 
 | |
6K1T
 
 | |
3C19
 
 | | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein MK0293 | | Authors: | Patskovsky, Y, Romero, R, Bonanno, J.B, Malashkevich, V, Dickey, M, Chang, S, Koss, J, Bain, K, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-22 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19.
To be Published
|
|
3C3M
 
 | | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1 | | Descriptor: | GLYCEROL, Response regulator receiver protein | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Meyer, A.J, Dickey, M, Chang, S, Groshong, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the N-terminal domain of response regulator receiver protein from Methanoculleus marisnigri JR1.
To be Published
|
|
3LSZ
 
 | | Crystal structure of glutathione s-transferase from Rhodobacter sphaeroides | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUTATHIONE, GLYCEROL, ... | | Authors: | Eswaramoorthy, S, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-02-14 | | Release date: | 2010-03-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of glutathione s-transferase from Rhodobacter sphaeroides
To be Published
|
|
6U07
 
 | | Computational Stabilization of T Cell Receptor Constant Domains | | Descriptor: | MAGNESIUM ION, Stabilized T cell receptor constant domain (Calpha), Stabilized T cell receptor constant domain (Cbeta) | | Authors: | Froning, K, Maguire, J, Sereno, A, Huang, F, Chang, S, Weichert, K, Frommelt, A.J, Dong, J, Wu, X, Austin, H, Conner, E.M, Fitchett, J.R, Heng, A.R, Balasubramaniam, D, Hilgers, M.T, Kuhlman, B, Demarest, S.J. | | Deposit date: | 2019-08-13 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Computational stabilization of T cell receptors allows pairing with antibodies to form bispecifics.
Nat Commun, 11, 2020
|
|
7NFX
 
 | | Mammalian ribosome nascent chain complex with SRP and SRP receptor in early state A | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Lee, J.H, Shan, S, Ban, N. | | Deposit date: | 2021-02-08 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Receptor compaction and GTPase rearrangement drive SRP-mediated cotranslational protein translocation into the ER.
Sci Adv, 7, 2021
|
|
1I4P
 
 | |
5IZ7
 
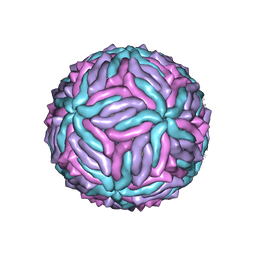 | | Cryo-EM structure of thermally stable Zika virus strain H/PF/2013 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, structural protein E, structural protein M | | Authors: | Kostyuchenko, V.A, Zhang, S, Fibriansah, G, Lok, S.M. | | Deposit date: | 2016-03-25 | | Release date: | 2016-05-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the thermally stable Zika virus
Nature, 533, 2016
|
|
5X9X
 
 | |
